Ligation sequencing gDNA - Native Barcoding Kit 24 V14 (SQK-NBD114.24) (NBE_9169_v114_revW_03Oct2025)
MinION: Protocol
Ligation sequencing gDNA - Native Barcoding Kit 24 V14 (SQK-NBD114.24) V NBE_9169_v114_revW_03Oct2025
This document describes the barcoding of native genomic DNA libraries.
The protocol:
- Requires the Native Barcoding Kit 24 V14 (SQK-NBD114.24)
- Is PCR-free
- Uses up to 24 barcodes
- Allows analysis of native DNA
- Is compatible with R10.4.1 flow cells
For Research Use Only
FOR RESEARCH USE ONLY
Contents
Introduction to the protocol
Library preparation
- 3. DNA repair and end-prep
- 4. Native barcode ligation
- 5. Adapter ligation and clean-up
- 6. Priming and loading the MinION and GridION Flow Cell
Sequencing and data analysis
Troubleshooting
Overview
This document describes the barcoding of native genomic DNA libraries.
The protocol:
- Requires the Native Barcoding Kit 24 V14 (SQK-NBD114.24)
- Is PCR-free
- Uses up to 24 barcodes
- Allows analysis of native DNA
- Is compatible with R10.4.1 flow cells
For Research Use Only
1. Overview of the protocol
Introduction to the Native Barcoding Kit 24 V14 protocol
This protocol describes how to carry out native barcoding of genomic DNA (gDNA) using the Native Barcoding Kit 24 V14 (SQK-NBD114.24). There are 24 unique barcodes available, allowing the user to pool up to 24 different samples in one sequencing experiment. It is highly recommended that a Lambda control experiment is completed first to become familiar with the technology.
Steps in the sequencing workflow:
Prepare for your experiment
You will need to:
- Extract your DNA, and check its length, quantity and purity. The quality checks performed during the protocol are essential in ensuring experimental success.
- Ensure you have your sequencing kit, the correct equipment and third-party reagents.
- Download the software for acquiring and analysing your data.
- Check your flow cell to ensure it has enough pores for a good sequencing run.
Prepare your library
The Table below is an overview of the steps required in the library preparation, including timings and stopping points.
| Library preparation | Process | Time | Stop option |
|---|---|---|---|
| DNA repair and end-prep | Repair the gDNA, and prepare the DNA ends for adapter attachment | 20 minutes | 4°C overnight |
| Native barcode ligation | Ligate the native barcodes to the DNA ends | 60 minutes | 4°C overnight |
| Adapter ligation and clean-up | Ligate sequencing adapters to the DNA ends | 50 minutes | 4°C for short-term storage or for repeated use, such as for reloading your flow cell –80°C for long-term storage |
| Priming and loading the flow cell | Prime the flow cell, and load your DNA library into the flow cell | 10 minutes |
Sequencing
You will need to:
- Start a sequencing run using the MinKNOW software, which will collect raw data from the device and convert it into basecalled reads.
- Demultiplex barcoded reads in MinKNOW, choosing the SQK-NBD114.24 kit option.
- Start the EPI2ME software and select a workflow for further analysis (this step is optional).
We do not recommend mixing barcoded libraries with non-barcoded libraries prior to sequencing.
Optional fragmentation and size selection
By default, the protocol contains no DNA fragmentation step, however in some cases it may be advantageous to fragment your sample. For example, when working with lower amounts of input gDNA (100 ng – 500 ng), fragmentation will increase the number of DNA molecules and therefore increase throughput. Instructions are available in the DNA Fragmentation section of Extraction methods.
Additionally, we offer several options for size-selecting your DNA sample to enrich for long fragments - instructions are available in the Size Selection section of Extraction methods.
Compatibility of this protocol
This protocol should only be used in combination with:
- Native Barcoding Kit 24 V14 (SQK-NBD114.24)
- R10.4.1 flow cells (FLO-MIN114)
- Flow Cell Wash Kit (EXP-WSH004)
- Sequencing Auxiliary Vials V14 (EXP-AUX003)
- Native Barcoding Expansion V14 (EXP-NBA114)
- MinION Mk1D - MinION Mk1D IT requirements document
- GridION - GridION IT requirements document
2. Equipment and consumables
Materials
- Native Barcoding Kit 24 V14 (SQK-NBD114.24)
- 400 ng gDNA per sample if using >4 barcodes
- OR 1000 ng gDNA per sample if using ≤4 barcodes
Consumables
- NEB Blunt/TA Ligase Master Mix (NEB, M0367)
- NEBNext FFPE Repair Mix (NEB, M6630)
- NEBNext Ultra II End repair/dA-tailing Module (NEB, E7546)
- NEBNext Quick Ligation Module (NEB, E6056)
- Eppendorf twin.tec® PCR plate 96 LoBind, semi-skirted (Merck, EP0030129504) with heat seals
- 1.5 ml Eppendorf DNA LoBind tubes
- 2 ml Eppendorf DNA LoBind tubes
- Nuclease-free water (e.g. Thermo Scientific, AM9937)
- Freshly prepared 80% ethanol in nuclease-free water
- Qubit™ Assay Tubes (Invitrogen, Q32856)
- Qubit™ dsDNA HS Assay Kit (ThermoFisher, Q32851)
- Bovine Serum Albumin (BSA) (50 mg/ml) (e.g Invitrogen™ UltraPure™ BSA 50 mg/ml, AM2616)
Equipment
- Hula mixer (gentle rotator mixer)
- Microplate centrifuge
- Microfuge
- Magnetic separation rack
- Vortex mixer
- Thermal cycler
- Multichannel pipette and tips
- P1000 pipette and tips
- P200 pipette and tips
- P100 pipette and tips
- P20 pipette and tips
- P10 pipette and tips
- P2 pipette and tips
- Ice bucket with ice
- Timer
- Eppendorf 5424 centrifuge (or equivalent)
- Qubit™ fluorometer (or equivalent for QC check)
Optional equipment
- Nanodrop spectrophotometer
For this protocol, we recommend the following inputs:
- 400 ng per sample for >4 barcodes
- 1000 ng per sample for ≤4 barcodes
Input DNA
How to QC your input DNA
It is important that the input DNA meets the quantity and quality requirements. Using too little or too much DNA, or DNA of poor quality (e.g. highly fragmented or containing RNA or chemical contaminants) can affect your library preparation.
For instructions on how to perform quality control of your DNA sample, please read the Input DNA/RNA QC protocol.
Chemical contaminants
Depending on how the DNA is extracted from the raw sample, certain chemical contaminants may remain in the purified DNA, which can affect library preparation efficiency and sequencing quality. Read more about contaminants on the Contaminants page of the Community.
Check your flow cell
We highly recommend that you check the number of pores in your flow cell prior to starting a sequencing experiment. This should be done within 12 weeks of purchasing your MinION/GridION/PromethION Flow Cells or within four weeks of purchasing Flongle Flow Cells. Oxford Nanopore Technologies will replace any unused flow cell with fewer than the number of pores listed in the Table below, when the result is reported within two days of performing the flow cell check, and when the storage recommendations have been followed. To do the flow cell check, please follow the instructions in the Flow Cell Check document.
| Flow cell | Minimum number of active pores covered by warranty |
|---|---|
| Flongle Flow Cell | 50 |
| MinION/GridION Flow Cell | 800 |
| PromethION Flow Cell | 5000 |
Third-party reagents
We have validated and recommend the use of all the third-party reagents used in this protocol. Alternatives have not been tested by Oxford Nanopore Technologies.
For all third-party reagents, we recommend following the manufacturer's instructions to prepare the reagents for use.
The Native Adapter (NA) used in this kit and protocol is not interchangeable with other sequencing adapters.
Native Barcoding Kit 24 V14 (SQK-NBD114.24) contents
Note: We are in the process of updating our native barcoding kits with an increased volume of Short Fragment Buffer (SFB). If you have an old format kit and/or require additional volume of Short Fragment Buffer (SFB), this can be purchased via our SFB Expansion (EXP-SFB001).
New format: increased volume of Short Fragment Buffer (SFB)
| Name | Acronym | Cap colour | No. of vials | Fill volume per vial (µl) |
|---|---|---|---|---|
| DNA Control Sample | DCS | Yellow | 2 | 35 |
| Native Adapter | NA | Green | 1 | 40 |
| Sequencing Buffer | SB | Red | 1 | 700 |
| Library Beads | LIB | Pink | 1 | 600 |
| Library Solution | LIS | White cap, pink label | 1 | 600 |
| Elution Buffer | EB | Black | 2 | 500 |
| AMPure XP Beads | AXP | Clear cap, light teal label | 1 | 6,000 |
| Long Fragment Buffer | LFB | Orange | 1 | 1,800 |
| Short Fragment Buffer | SFB | Clear | 1 | 13,000 |
| EDTA | EDTA | Blue | 1 | 700 |
| Flow Cell Flush | FCF | Clear cap, light blue label | 1 | 8,000 |
| Flow Cell Tether | FCT | Purple | 1 | 200 |
| Native Barcode plate | NB01-24 | - | 2 plates, 3 sets of barcodes per plate | 5 µl per well |
Old format: lower volume of Short Fragment Buffer (SFB)
| Name | Acronym | Cap colour | No. of vials | Fill volume per vial (µl) |
|---|---|---|---|---|
| DNA Control Sample | DCS | Yellow | 2 | 35 |
| Native Adapter | NA | Green | 1 | 40 |
| Sequencing Buffer | SB | Red | 1 | 700 |
| Library Beads | LIB | Pink | 1 | 600 |
| Library Solution | LIS | White cap, pink label | 1 | 600 |
| Elution Buffer | EB | Black | 2 | 500 |
| AMPure XP Beads | AXP | Clear cap, light teal label | 1 | 6,000 |
| Long Fragment Buffer | LFB | Orange | 1 | 1,800 |
| Short Fragment Buffer | SFB | Clear | 1 | 1,800 |
| EDTA | EDTA | Blue | 1 | 700 |
| Flow Cell Flush | FCF | Clear cap, light blue label | 1 | 8,000 |
| Flow Cell Tether | FCT | Purple | 1 | 200 |
| Native Barcode plate | NB01-24 | - | 2 plates, 3 sets of barcodes per plate | 5 µl per well |
Note: This product contains AMPure XP reagent manufactured by Beckman Coulter, Inc. and can be stored at -20°C with the kit without detriment to reagent stability.
Note: The DNA Control Sample (DCS) is a 3.6 kb standard amplicon mapping the 3' end of the Lambda genome.
To maximise the use of the Native Barcoding Kits, the Native Barcode Auxiliary V14 (EXP-NBA114) and the Sequencing Auxiliary Vials V14 (EXP-AUX003) expansion packs are available.
These expansions provide extra library preparation and flow cell priming reagents to allow users to utilise any unused barcodes for those running in smaller subsets.
Both expansion packs used together will provide enough reagents for 12 reactions. For customers requiring extra EDTA to maximise the use of barcodes, we recommend using 0.25 M EDTA and adding 4 µl for library preps using the SQK-NBD114.24 kit and 2 µl for preps using the SQK-NBD114.96 kit.
Native Barcode Auxiliary V14 (EXP-NBA114) contents:
Note: This product contains AMPure XP reagent manufactured by Beckman Coulter, Inc. and can be stored at -20°C with the kit without detriment to reagent stability.
Sequencing Auxiliary Vials V14 (EXP-AUX003) contents:
Native barcode sequences
| Component | Forward sequence | Reverse sequence |
|---|---|---|
| NB01 | CACAAAGACACCGACAACTTTCTT | AAGAAAGTTGTCGGTGTCTTTGTG |
| NB02 | ACAGACGACTACAAACGGAATCGA | TCGATTCCGTTTGTAGTCGTCTGT |
| NB03 | CCTGGTAACTGGGACACAAGACTC | GAGTCTTGTGTCCCAGTTACCAGG |
| NB04 | TAGGGAAACACGATAGAATCCGAA | TTCGGATTCTATCGTGTTTCCCTA |
| NB05 | AAGGTTACACAAACCCTGGACAAG | CTTGTCCAGGGTTTGTGTAACCTT |
| NB06 | GACTACTTTCTGCCTTTGCGAGAA | TTCTCGCAAAGGCAGAAAGTAGTC |
| NB07 | AAGGATTCATTCCCACGGTAACAC | GTGTTACCGTGGGAATGAATCCTT |
| NB08 | ACGTAACTTGGTTTGTTCCCTGAA | TTCAGGGAACAAACCAAGTTACGT |
| NB09 | AACCAAGACTCGCTGTGCCTAGTT | AACTAGGCACAGCGAGTCTTGGTT |
| NB10 | GAGAGGACAAAGGTTTCAACGCTT | AAGCGTTGAAACCTTTGTCCTCTC |
| NB11 | TCCATTCCCTCCGATAGATGAAAC | GTTTCATCTATCGGAGGGAATGGA |
| NB12 | TCCGATTCTGCTTCTTTCTACCTG | CAGGTAGAAAGAAGCAGAATCGGA |
| NB13 | AGAACGACTTCCATACTCGTGTGA | TCACACGAGTATGGAAGTCGTTCT |
| NB14 | AACGAGTCTCTTGGGACCCATAGA | TCTATGGGTCCCAAGAGACTCGTT |
| NB15 | AGGTCTACCTCGCTAACACCACTG | CAGTGGTGTTAGCGAGGTAGACCT |
| NB16 | CGTCAACTGACAGTGGTTCGTACT | AGTACGAACCACTGTCAGTTGACG |
| NB17 | ACCCTCCAGGAAAGTACCTCTGAT | ATCAGAGGTACTTTCCTGGAGGGT |
| NB18 | CCAAACCCAACAACCTAGATAGGC | GCCTATCTAGGTTGTTGGGTTTGG |
| NB19 | GTTCCTCGTGCAGTGTCAAGAGAT | ATCTCTTGACACTGCACGAGGAAC |
| NB20 | TTGCGTCCTGTTACGAGAACTCAT | ATGAGTTCTCGTAACAGGACGCAA |
| NB21 | GAGCCTCTCATTGTCCGTTCTCTA | TAGAGAACGGACAATGAGAGGCTC |
| NB22 | ACCACTGCCATGTATCAAAGTACG | CGTACTTTGATACATGGCAGTGGT |
| NB23 | CTTACTACCCAGTGAACCTCCTCG | CGAGGAGGTTCACTGGGTAGTAAG |
| NB24 | GCATAGTTCTGCATGATGGGTTAG | CTAACCCATCATGCAGAACTATGC |
3. DNA repair and end-prep
Materials
- 400 ng gDNA per barcode
- OR 1000 ng gDNA per sample if using ≤4 barcodes
- AMPure XP Beads (AXP)
- DNA Control Sample (DCS)
Consumables
- NEBNext® FFPE DNA Repair Mix (NEB, M6630)
- NEBNext® Ultra™ II End Repair/dA-Tailing Module (NEB, E7546)
- Freshly prepared 80% ethanol in nuclease-free water
- 1.5 ml Eppendorf DNA LoBind tubes
- Eppendorf twin.tec® PCR plate 96 LoBind, semi-skirted (Merck, EP0030129504) with heat seals
- OR 0.2 ml thin-walled PCR tubes
- Nuclease-free water (e.g. Thermo Scientific, AM9937)
- Qubit™ Assay Tubes (Invitrogen, Q32856)
- Qubit dsDNA HS Assay Kit (Invitrogen, Q32851)
Equipment
- P1000 pipette and tips
- P200 pipette and tips
- P100 pipette and tips
- P20 pipette and tips
- P10 pipette and tips
- P2 pipette and tips
- Multichannel pipette and tips
- Thermal cycler
- Microplate centrifuge
- Microfuge
- Ice bucket with ice
- Magnetic separation rack
- Vortex mixer
- Hula mixer (rotator mixer)
- Qubit fluorometer (or equivalent)
For samples containing long gDNA fragments, we recommend using wide-bore pipette tips for the mixing steps to preserve the DNA length.
Thaw the AMPure XP Beads (AXP) and DNA Control Sample (DCS) at room temperature and mix by vortexing. Keep the beads at room temperature and store the DNA Control Sample (DCS) on ice.
Prepare the NEBNext FFPE DNA Repair Mix and NEBNext Ultra II End Repair / dA-tailing Module reagents in accordance with manufacturer’s instructions, and place on ice.
For optimal performance, NEB recommend the following:
Thaw all reagents on ice.
Flick and/or invert the reagent tubes to ensure they are well mixed.
Note: Do not vortex the FFPE DNA Repair Mix or Ultra II End Prep Enzyme Mix.Always spin down tubes before opening for the first time each day.
The Ultra II End Prep Reaction Buffer and FFPE DNA Repair Buffer may have a little precipitate. Allow the mixtures to come to room temperature and pipette the buffer up and down several times to break up the precipitate, followed by vortexing the tube for 30 seconds to solubilise any precipitate.
Note: It is important the buffers are mixed well by vortexing.The FFPE DNA Repair Buffer may have a yellow tinge and is fine to use if yellow.
Do not vortex the NEBNext FFPE DNA Repair Mix or NEBNext Ultra II End Prep Enzyme Mix.
It is important that the NEBNext FFPE DNA Repair Buffer and NEBNext Ultra II End Prep Reaction Buffer are mixed well by vortexing.
Check for any visible precipitate; vortexing for at least 30 seconds may be required to solubilise any precipitate.
Dilute your DNA Control Sample (DCS) by adding 105 µl Elution Buffer (EB) directly to one DCS tube. Mix gently by pipetting and spin down.
One tube of diluted DNA Control Sample (DCS) is enough for 140 samples. Excess can be stored at -20°C in the freezer.
We recommend using the DNA Control Sample (DCS) in your library prep for troubleshooting purposes. However, you can omit this step and make up the extra 1 µl with your sample DNA.
In clean 0.2 ml thin-walled PCR tubes (or a clean 96-well plate), prepare your DNA samples:
- For >4 barcodes, aliquot 400 ng per sample
- For ≤4 barcodes, aliquot 1000 ng per sample
Make up each sample to 11 µl using nuclease-free water. Mix gently by pipetting and spin down.
Combine the following components per tube/well:
Between each addition, pipette mix 10-20 times.
| Reagent | Volume |
|---|---|
| DNA sample | 11 µl |
| Diluted DNA Control Sample (DCS) | 1 µl |
| NEBNext FFPE DNA Repair Buffer | 0.875 µl |
| Ultra II End-prep Reaction Buffer | 0.875 µl |
| Ultra II End-prep Enzyme Mix | 0.75 µl |
| NEBNext FFPE DNA Repair Mix | 0.5 µl |
| Total | 15 µl |
We recommend making up a mastermix of the End Prep and DNA Repair reagents for the total number of samples and adding 3 µl to each well.
Ensure the components are thoroughly mixed by pipetting and spin down in a centrifuge.
Using a thermal cycler, incubate at 20°C for 5 minutes and 65°C for 5 minutes.
Transfer each sample into a clean 1.5 ml Eppendorf DNA LoBind tube.
Resuspend the AMPure XP beads (AXP) by vortexing.
Add 15 µl of resuspended AMPure XP Beads (AXP) to each end-prep reaction and mix by flicking the tube.
Incubate on a Hula mixer (rotator mixer) for 5 minutes at room temperature.
Prepare sufficient fresh 80% ethanol in nuclease-free water for all of your samples. Allow enough for 400 µl per sample, with some excess.
Spin down the samples and pellet the beads on a magnet until the eluate is clear and colourless. Keep the tubes on the magnet and pipette off the supernatant.
Keep the tube on the magnet and wash the beads with 200 µl of freshly prepared 80% ethanol without disturbing the pellet. Remove the ethanol using a pipette and discard.
If the pellet was disturbed, wait for beads to pellet again before removing the ethanol.
Repeat the previous step.
Briefly spin down and place the tubes back on the magnet for the beads to pellet. Pipette off any residual ethanol. Allow to dry for 30 seconds, but do not dry the pellets to the point of cracking.
Remove the tubes from the magnetic rack and resuspend the pellet in 10 µl nuclease-free water. Spin down and incubate for 2 minutes at room temperature.
Pellet the beads on a magnet until the eluate is clear and colourless.
Remove and retain 10 µl of eluate into a clean 1.5 ml Eppendorf DNA LoBind tube.
- Dispose of the pelleted beads
Quantify 1 µl of each eluted sample using a Qubit fluorometer.
Take forward an equimolar mass of each sample to be barcoded forward into the native barcode ligation step. However, you may store the samples at 4°C overnight.
4. Native barcode ligation
Materials
- Native Barcodes (NB01-24)
- AMPure XP Beads (AXP)
- EDTA (EDTA)
- S Fragment Buffer (SFB)
Consumables
- NEB Blunt/TA Ligase Master Mix (NEB, M0367)
- Nuclease-free water (e.g. Thermo Scientific, AM9937)
- 1.5 ml Eppendorf DNA LoBind tubes
- Eppendorf twin.tec® PCR plate 96 LoBind, semi-skirted (Merck, EP0030129504) with heat seals
- OR 0.2 ml thin-walled PCR tubes
- Qubit™ Assay Tubes (Invitrogen, Q32856)
- Qubit™ dsDNA HS Assay Kit (ThermoFisher, Q32851)
Equipment
- Magnetic separation rack
- Vortex mixer
- Hula mixer (gentle rotator mixer)
- Microfuge
- Thermal cycler
- Ice bucket with ice
- Multichannel pipette and tips
- P1000 pipette and tips
- P200 pipette and tips
- P100 pipette and tips
- P20 pipette and tips
- P10 pipette and tips
- P2 pipette and tips
- Qubit™ fluorometer (or equivalent for QC check)
Prepare the NEB Blunt/TA Ligase Master Mix according to the manufacturer's instructions, and place on ice:
Thaw the reagent at room temperature.
Spin down the reagent tube for 5 seconds.
Ensure the reagent is fully mixed by performing 10 full volume pipette mixes.
Thaw the EDTA at room temperature and mix by vortexing. Then spin down and place on ice.
Thaw the Short Fragment Buffer (SFB) at room temperature and mix by vortexing. Place on ice.
Thaw the Native Barcodes (NB01-24) at room temperature. Briefly spin down, individually mix the barcodes required for your number of samples by pipetting, and place them on ice.
The wells of the barcoding plate are intended for single use only. Please ensure your barcode well is sealed before use, and do not reuse the barcode well once pierced/opened.
Select a unique barcode for each sample to be run together on the same flow cell. Up to 24 samples can be barcoded and combined in one experiment.
Please note: Only use one barcode per sample.
In clean 0.2 ml PCR-tubes or a 96-well plate, add the reagents in the following order per well:
Between each addition, pipette mix 10-20 times.
| Reagent | Volume |
|---|---|
| End-prepped DNA | 7.5 µl |
| Native Barcode (NB01-24) | 2.5 µl |
| Blunt/TA Ligase Master Mix | 10 µl |
| Total | 20 µl |
Thoroughly mix the reaction by gently pipetting and briefly spinning down.
Incubate for 20 minutes at room temperature.
Add 4 µl EDTA (blue cap) to each well and mix thoroughly by pipetting and spin down briefly.
EDTA is added at this step to stop the reaction.
Pool all the barcoded samples in a 1.5 ml Eppendorf DNA LoBind tube.
| Volume per sample | For 6 samples | For 12 samples | For 24 samples | |
|---|---|---|---|---|
| Total volume for preps using EDTA (blue cap) | 24 µl | 144 µl | 288 µl | 576 µl |
We recommend checking the base of your tubes/plate are all the same volume before pooling and after to ensure all the liquid has been taken forward.
Resuspend the AMPure XP Beads (AXP) by vortexing.
Add 0.4X AMPure XP Beads (AXP) to the pooled reaction, and mix by pipetting.
| Volume per sample | For 6 samples | For 12 samples | For 24 samples | |
|---|---|---|---|---|
| Volume of AXP for preps using EDTA (blue cap) | 10 µl | 58 µl | 115 µl | 230 µl |
Incubate on a Hula mixer (rotator mixer) for 10 minutes at room temperature.
For optimal results we recommend using Short Fragment Buffer (SFB) for the clean-up steps following native barcoding.
Our development teams have determined that using the Short Fragment Buffer (SFB) instead of ethanol for the post-barcoding washes removes excess barcode more efficiently. This translates to improved barcode classification rates and reduced physical barcode cross-talk.
New batches of the native barcoding kits will contain sufficient Short Fragment Buffer (SFB) to follow the updated method. If you have an older format of the native barcoding kit with a lower volume of Short Fragment Buffer (SFB), you may require additional reagents available through the SFB Expansion (EXP-SFB001).
Please note, the old method using 80% ethanol is still compatible with this method. If you wish to continue using 80% ethanol for your post-barcoding wash, please follow the steps below:
- Prepare sufficient fresh 80% ethanol in nuclease-free water for your washes.
- Use the freshly prepared 80% ethanol in place of the Short Fragment Buffer (SFB) for the wash steps below.
Spin down the sample and pellet on a magnet for 5 minutes. Keep the tube on the magnetic rack until the eluate is clear and colourless, and pipette off the supernatant.
Wash the beads with 700 µl of Short Fragment Buffer (SFB). Flick the beads to resuspend, spin down, then return the sample to the magnetic rack and allow the beads to pellet. Remove the buffer using a pipette and discard.
Repeat the previous step.
Spin down and place the tube back on the magnetic rack. Pipette off any residual buffer.
Remove the tube from the magnetic rack and resuspend the pellet in 35 µl nuclease-free water by gently flicking.
Incubate for 10 minutes at 37°C. Every 2 minutes, agitate the sample by gently flicking for 10 seconds to encourage DNA elution.
Pellet the beads on a magnetic rack until the eluate is clear and colourless.
Remove and retain 35 µl of eluate into a clean 1.5 ml Eppendorf DNA LoBind tube.
Quantify 1 µl of eluted sample using a Qubit fluorometer.
Take forward the barcoded DNA library to the adapter ligation and clean-up step. However, you may store the sample at 4°C overnight.
5. Adapter ligation and clean-up
Materials
- Long Fragment Buffer (LFB)
- S Fragment Buffer (SFB)
- Elution Buffer (EB)
- Native Adapter (NA)
- AMPure XP Beads (AXP)
Consumables
- NEBNext® Quick Ligation Module (NEB, E6056)
- 1.5 ml Eppendorf DNA LoBind tubes
- Qubit™ Assay Tubes (Invitrogen, Q32856)
- Qubit™ dsDNA HS Assay Kit (ThermoFisher, Q32851)
Equipment
- Microfuge
- Magnetic separation rack
- Vortex mixer
- Hula mixer (gentle rotator mixer)
- Thermal cycler
- P1000 pipette and tips
- P200 pipette and tips
- P100 pipette and tips
- P20 pipette and tips
- P10 pipette and tips
- Ice bucket with ice
- Qubit™ fluorometer (or equivalent for QC check)
The Native Adapter (NA) used in this kit and protocol is not interchangeable with other sequencing adapters.
Check your flow cell.
We recommend performing a flow cell check before starting adapter ligation and clean-up to ensure you have a flow cell with sufficient pores for a good sequencing run.
See the flow cell check document for more information.
Prepare the NEBNext Quick Ligation Reaction Module according to the manufacturer's instructions, and place on ice:
Thaw the reagents at room temperature.
Spin down the reagent tubes for 5 seconds.
Ensure the reagents are fully mixed by performing 10 full volume pipette mixes. Note: Do NOT vortex the Quick T4 DNA Ligase.
The NEBNext Quick Ligation Reaction Buffer (5x) may have a little precipitate. Allow the mixture to come to room temperature and pipette the buffer up and down several times to break up the precipitate, followed by vortexing the tube for several seconds to ensure the reagent is thoroughly mixed.
Do not vortex the Quick T4 DNA Ligase.
Spin down the Native Adapter (NA) and Quick T4 DNA Ligase, pipette mix and place on ice.
Thaw the Elution Buffer (EB) at room temperature and mix by vortexing. Then spin down and place on ice.
Depending on the wash buffer (LFB or SFB) used, the clean-up step after adapter ligation is designed to either enrich for DNA fragments of >3 kb, or purify all fragments equally.
- To enrich for DNA fragments of 3 kb or longer, use Long Fragment Buffer (LFB).
- To retain DNA fragments of all sizes, use Short Fragment Buffer (SFB).
Thaw either Long Fragment Buffer (LFB) or Short Fragment Buffer (SFB) at room temperature and mix by vortexing. Then spin down and keep at room temperature.
In a 1.5 ml Eppendorf LoBind tube, mix in the following order:
Between each addition, pipette mix 10-20 times.
| Reagent | Volume |
|---|---|
| Pooled barcoded sample | 30 µl |
| Native Adapter (NA) | 5 µl |
| NEBNext Quick Ligation Reaction Buffer (5X) | 10 µl |
| Quick T4 DNA Ligase | 5 µl |
| Total | 50 µl |
Thoroughly mix the reaction by gently pipetting and briefly spinning down.
Incubate the reaction for 20 minutes at room temperature.
The next clean-up step uses Long Fragment Buffer (LFB) or Short Fragment Buffer (SFB) rather than 80% ethanol to wash the beads. The use of ethanol will be detrimental to the sequencing reaction.
Resuspend the AMPure XP Beads (AXP) by vortexing.
Add 20 µl of resuspended AMPure XP Beads (AXP) to the reaction and mix by pipetting.
Incubate on a Hula mixer (rotator mixer) for 10 minutes at room temperature.
Spin down the sample and pellet on the magnetic rack. Keep the tube on the magnet and pipette off the supernatant.
Wash the beads by adding either 125 μl Long Fragment Buffer (LFB) or Short Fragment Buffer (SFB). Flick the beads to resuspend, spin down, then return the tube to the magnetic rack and allow the beads to pellet. Remove the supernatant using a pipette and discard.
Repeat the previous step.
Spin down and place the tube back on the magnet. Pipette off any residual supernatant.
Remove the tube from the magnetic rack and resuspend pellet in 15 µl Elution Buffer (EB).
Spin down and incubate for 10 minutes at 37°C. Every 2 minutes, agitate the sample by gently flicking for 10 seconds to encourage DNA elution.
Pellet the beads on a magnet until the eluate is clear and colourless, for at least 1 minute.
Remove and retain 15 µl of eluate containing the DNA library into a clean 1.5 ml Eppendorf DNA LoBind tube.
Dispose of the pelleted beads
Quantify 1 µl of eluted sample using a Qubit fluorometer.
Depending on your DNA library fragment size, prepare your final library in 12 µl of Elution Buffer (EB).
| Fragment library length | Flow cell loading amount |
|---|---|
| Very short (<1 kb) | 100 fmol |
| Short (1-10 kb) | 35–50 fmol |
| Long (>10 kb) | 300 ng |
Note: If the library yields are below the input recommendations, load the entire library.
If required, we recommend using a mass to mol calculator such as the NEB calculator.
The prepared library is used for loading onto the flow cell. Store the library on ice or at 4°C until ready to load.
Library storage recommendations
We recommend storing libraries in Eppendorf DNA LoBind tubes at 4°C for short-term storage or repeated use, for example, re-loading flow cells between washes. For single use and long-term storage of more than 3 months, we recommend storing libraries at -80°C in Eppendorf DNA LoBind tubes.
If quantities allow, the library may be diluted in Elution Buffer (EB) for splitting across multiple flow cells.
Depending on how many flow cells the library will be split across, more Elution Buffer (EB) than what is supplied in the kit will be required.
6. Priming and loading the MinION and GridION Flow Cell
Materials
- Flow Cell Flush (FCF)
- Flow Cell Tether (FCT)
- Library Solution (LIS)
- Library Beads (LIB)
- Sequencing Buffer (SB)
Consumables
- 1.5 ml Eppendorf DNA LoBind tubes
- MinION/GridION Flow Cell
- Nuclease-free water (e.g. Thermo Scientific, AM9937)
- Bovine Serum Albumin (BSA) (50 mg/ml) (e.g Invitrogen™ UltraPure™ BSA 50 mg/ml, AM2616)
Equipment
- MinION or GridION device
- MinION/GridION Flow Cell Light Shield
- P1000 pipette and tips
- P100 pipette and tips
- P20 pipette and tips
- P10 pipette and tips
Please note, this kit is only compatible with R10.4.1 flow cells (FLO-MIN114).
Take the flow cell out of the fridge and leave it at room temperature for 20 minutes. This will improve visibility of the array during priming and sample loading.
Priming and loading a flow cell
We recommend all new users watch the 'Priming and loading your flow cell' video before your first run.
Using the Library Solution
For most sequencing experiments, use the Library Beads (LIB) for loading your library onto the flow cell. However, for viscous libraries it may be difficult to load with the beads and may be appropriate to load using the Library Solution (LIS).
Thaw the Sequencing Buffer (SB), Library Beads (LIB) or Library Solution (LIS, if using), Flow Cell Tether (FCT) and Flow Cell Flush (FCF) at room temperature before mixing by vortexing. Then spin down and store on ice.
For optimal sequencing performance and improved output on MinION R10.4.1 flow cells (FLO-MIN114), add Bovine Serum Albumin (BSA) to the flow cell priming mix at a final concentration of 0.2 mg/ml.
Note: We do not recommend using any other albumin type (e.g. recombinant human serum albumin).
To prepare the flow cell priming mix with BSA, combine Flow Cell Flush (FCF) and Flow Cell Tether (FCT), as directed below. Mix by pipetting at room temperature.
In a suitable tube for the number of flow cells, combine the following reagents:
| Reagent | Volume per flow cell |
|---|---|
| Flow Cell Flush (FCF) | 1,170 µl |
| Bovine Serum Albumin (BSA) at 50 mg/ml | 5 µl |
| Flow Cell Tether (FCT) | 30 µl |
| Total volume | 1,205 µl |
Open the MinION or GridION device lid and slide the flow cell under the clip. Press down firmly on the priming port cover to ensure correct thermal and electrical contact.
Complete a flow cell check to assess the number of pores available before loading the library.
This step can be omitted if the flow cell has been checked previously.
See the flow cell check document for more information.
Slide the flow cell priming port cover clockwise to open the priming port.
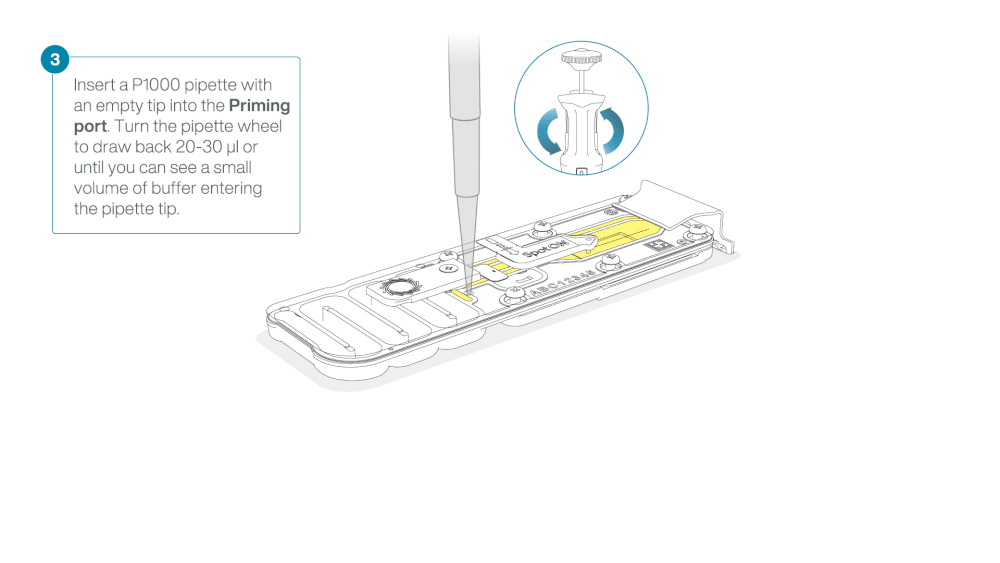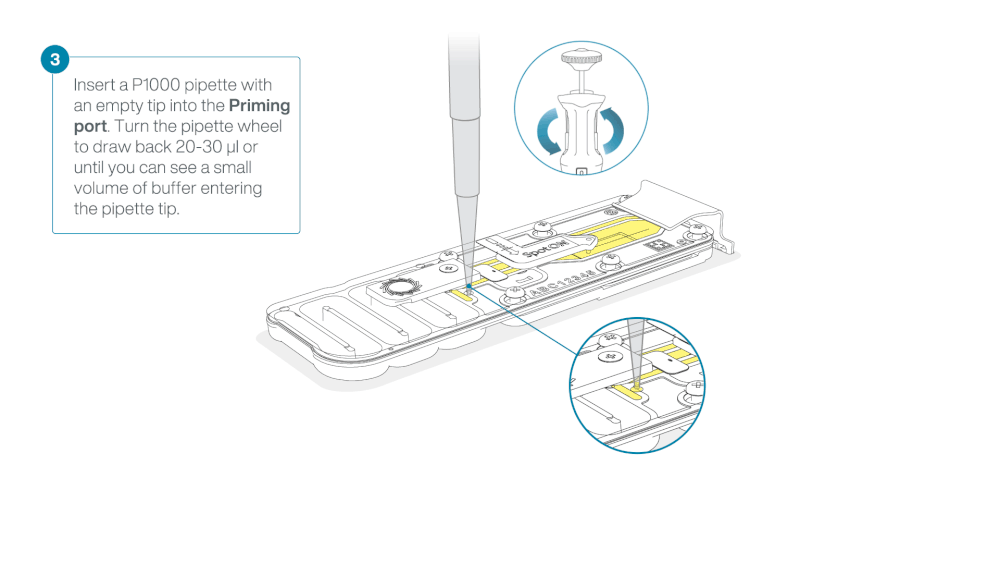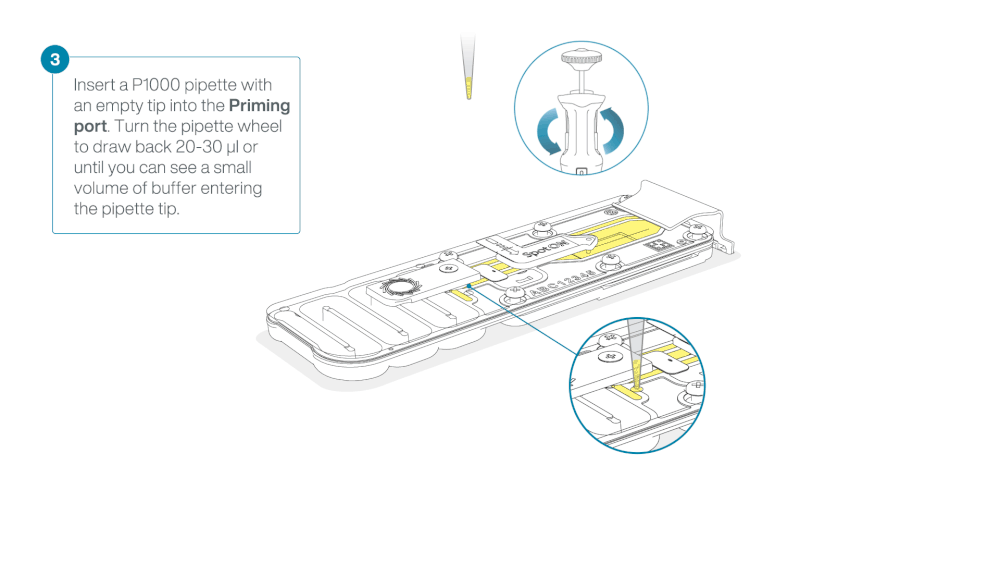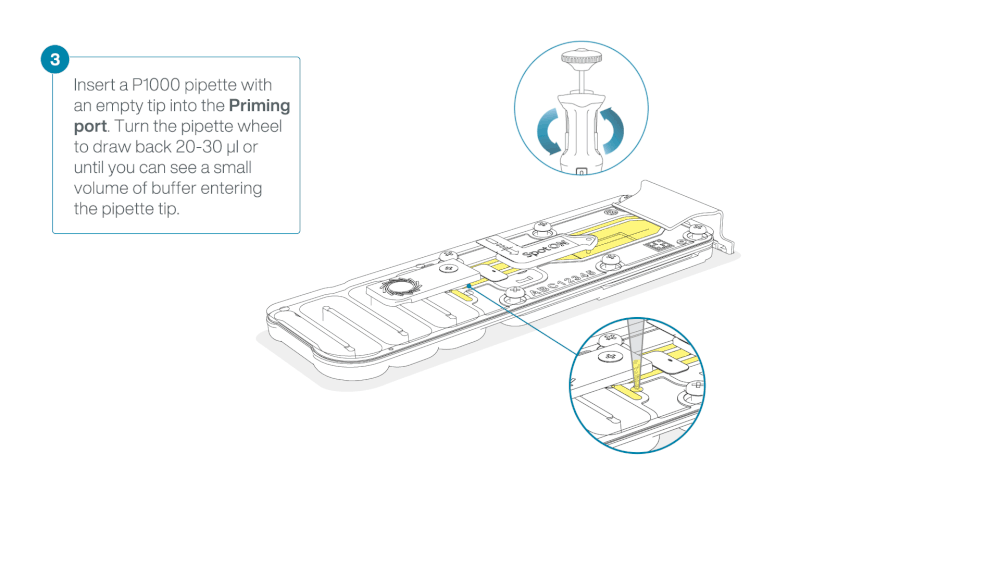
Take care when drawing back buffer from the flow cell. Do not remove more than 20-30 µl, and make sure that the array of pores are covered by buffer at all times. Introducing air bubbles into the array can irreversibly damage pores.
After opening the priming port, check for a small air bubble under the cover. Draw back a small volume to remove any bubbles:
- Set a P1000 pipette to 200 µl
- Insert the tip into the priming port
- Turn the wheel until the dial shows 220-230 µl, to draw back 20-30 µl, or until you can see a small volume of buffer entering the pipette tip
Note: Visually check that there is continuous buffer from the priming port across the sensor array.
Load 800 µl of the priming mix into the flow cell via the priming port, avoiding the introduction of air bubbles. Wait for five minutes. During this time, prepare the library for loading by following the steps below.

Thoroughly mix the contents of the Library Beads (LIB) by pipetting.
The Library Beads (LIB) tube contains a suspension of beads. These beads settle very quickly. It is vital that they are mixed immediately before use.
We recommend using the Library Beads (LIB) for most sequencing experiments. However, the Library Solution (LIS) is available for more viscous libraries.
In a new 1.5 ml Eppendorf DNA LoBind tube, prepare the library for loading as follows:
| Reagent | Volume per flow cell |
|---|---|
| Sequencing Buffer (SB) | 37.5 µl |
| Library Beads (LIB) mixed immediately before use, or Library Solution (LIS), if using | 25.5 µl |
| DNA library | 12 µl |
| Total | 75 µl |
Complete the flow cell priming:
- Gently lift the SpotON sample port cover to make the SpotON sample port accessible.
- Load 200 µl of the priming mix into the flow cell priming port (not the SpotON sample port), avoiding the introduction of air bubbles.

Mix the prepared library gently by pipetting up and down just prior to loading.
Add 75 μl of the prepared library to the flow cell via the SpotON sample port in a dropwise fashion. Ensure each drop flows into the port before adding the next.

Gently replace the SpotON sample port cover, making sure the bung enters the SpotON port and close the priming port.

For optimal sequencing output, install the light shield on your flow cell as soon as the library has been loaded.
We recommend leaving the light shield on the flow cell when library is loaded, including during any washing and reloading steps. The shield can be removed when the library has been removed from the flow cell.
Place the light shield onto the flow cell, as follows:
Carefully place the leading edge of the light shield against the clip. Note: Do not force the light shield underneath the clip.
Gently lower the light shield onto the flow cell. The light shield should sit around the SpotON cover, covering the entire top section of the flow cell.

The MinION Flow Cell Light Shield is not secured to the flow cell and careful handling is required after installation.
Close the device lid and set up a sequencing run on MinKNOW.
When a flow cell is inserted into the MinION Mk1D, the device lid will sit on top of the flow cell, leaving a small gap around the sides. This is normal and has no impact on the performance of the device.
Please refer to this FAQ regarding the device lid.

7. Data acquisition and basecalling
How to start sequencing
Once you have loaded your flow cell, the sequencing run can be started on MinKNOW, our sequencing software that controls the device, data acquisition and real-time basecalling. For more detailed information on setting up and using MinKNOW, please see the MinKNOW protocol.
MinKNOW can be used and set up to sequence in multiple ways:
- On a computer either directly or remotely connected to a sequencing device.
- Directly on a GridION sequencing device.
For more information on using MinKNOW on a sequencing device, please see the device user manuals:
To start a sequencing run on MinKNOW:
1. Navigate to the start page and click Start sequencing.
2. Fill in your experiment details, such as name and flow cell position and sample ID.
3. Select the Native Barcoding Kit 24 V14 (SQK-NBD114.24) on the Kit page.
4. Configure the sequencing and output parameters for your sequencing run or keep to the default settings on the Run configuration tab.
Note: If basecalling was turned off when a sequencing run was set up, basecalling can be performed post-run on MinKNOW. For more information, please see the MinKNOW protocol.
5. Click Start to initiate the sequencing run.
Data analysis after sequencing
After sequencing has completed on MinKNOW, the flow cell can be reused or returned, as outlined in the Flow cell reuse and returns section.
After sequencing and basecalling, the data can be analysed. For further information about options for basecalling and post-basecalling analysis, please refer to the Data Analysis document.
In the Downstream analysis section, we outline further options for analysing your data.
8. Flow cell reuse and returns
Materials
- Flow Cell Wash Kit (EXP-WSH004)
After your sequencing experiment is complete, if you would like to reuse the flow cell, please follow the Flow Cell Wash Kit protocol and store the washed flow cell at +2°C to +8°C.
The Flow Cell Wash Kit protocol is available on the Nanopore Community.
We recommend you to wash the flow cell as soon as possible after you stop the run. However, if this is not possible, leave the flow cell on the device and wash it the next day.
Alternatively, follow the returns procedure to send the flow cell back to Oxford Nanopore.
Instructions for returning flow cells can be found here.
If you encounter issues or have questions about your sequencing experiment, please refer to the Troubleshooting Guide that can be found in this protocol.
9. Downstream analysis
Post-basecalling analysis
There are several options for further analysing your basecalled data:
EPI2ME workflows
For in-depth data analysis, Oxford Nanopore Technologies offers a range of bioinformatics tutorials and workflows available in EPI2ME, which are available in the EPI2ME section of the Community. The platform provides a vehicle where workflows deposited in GitHub by our Research and Applications teams can be showcased with descriptive texts, functional bioinformatics code and example data.
Research analysis tools
Oxford Nanopore Technologies' Research division has created a number of analysis tools, that are available in the Oxford Nanopore GitHub repository. The tools are aimed at advanced users, and contain instructions for how to install and run the software. They are provided as-is, with minimal support.
Community-developed analysis tools
If a data analysis method for your research question is not provided in any of the resources above, please refer to the resource centre and search for bioinformatics tools for your application. Numerous members of the Nanopore Community have developed their own tools and pipelines for analysing nanopore sequencing data, most of which are available on GitHub. Please be aware that these tools are not supported by Oxford Nanopore Technologies, and are not guaranteed to be compatible with the latest chemistry/software configuration.
10. Issues during DNA/RNA extraction and library preparation for Kit 14
Below is a list of the most commonly encountered issues, with some suggested causes and solutions.
We also have an FAQ section available on the Nanopore Community Support section.
If you have tried our suggested solutions and the issue still persists, please contact Technical Support via email (support@nanoporetech.com) or via LiveChat in the Nanopore Community.
Low sample quality
| Observation | Possible cause | Comments and actions |
|---|---|---|
| Low DNA purity (Nanodrop reading for DNA OD 260/280 is <1.8 and OD 260/230 is <2.0–2.2) | The DNA extraction method does not provide the required purity | The effects of contaminants are shown in the Contaminants document. Please try an alternative extraction method that does not result in contaminant carryover. Consider performing an additional SPRI clean-up step. |
| Low RNA integrity (RNA integrity number <9.5 RIN, or the rRNA band is shown as a smear on the gel) | The RNA degraded during extraction | Try a different RNA extraction method. For more info on RIN, please see the RNA Integrity Number document. Further information can be found in the DNA/RNA Handling page. |
| RNA has a shorter than expected fragment length | The RNA degraded during extraction | Try a different RNA extraction method. For more info on RIN, please see the RNA Integrity Number document. Further information can be found in the DNA/RNA Handling page. We recommend working in an RNase-free environment, and to keep your lab equipment RNase-free when working with RNA. |
Low DNA recovery after AMPure bead clean-up
| Observation | Possible cause | Comments and actions |
|---|---|---|
| Low recovery | DNA loss due to a lower than intended AMPure beads-to-sample ratio | 1. AMPure beads settle quickly, so ensure they are well resuspended before adding them to the sample. 2. When the AMPure beads-to-sample ratio is lower than 0.4:1, DNA fragments of any size will be lost during the clean-up. |
| Low recovery | DNA fragments are shorter than expected | The lower the AMPure beads-to-sample ratio, the more stringent the selection against short fragments. Please always determine the input DNA length on an agarose gel (or other gel electrophoresis methods) and then calculate the appropriate amount of AMPure beads to use. 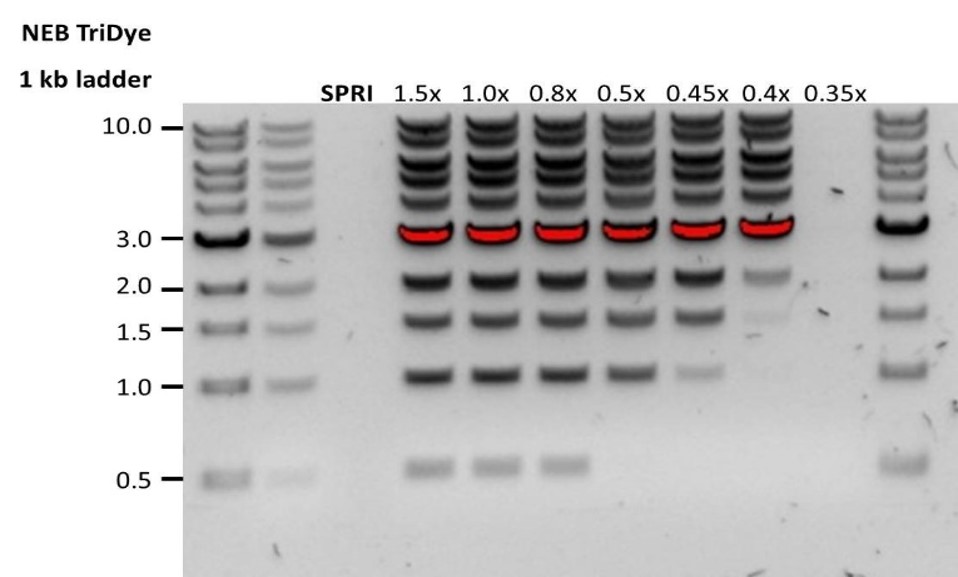 |
| Low recovery after end-prep | The wash step used ethanol <70% | DNA will be eluted from the beads when using ethanol <70%. Make sure to use the correct percentage. |
11. Issues during the sequencing run for Kit 14
Below is a list of the most commonly encountered issues, with some suggested causes and solutions.
We also have an FAQ section available on the Nanopore Community Support section.
If you have tried our suggested solutions and the issue still persists, please contact Technical Support via email (support@nanoporetech.com) or via LiveChat in the Nanopore Community.
Fewer pores at the start of sequencing than after Flow Cell Check
| Observation | Possible cause | Comments and actions |
|---|---|---|
| MinKNOW reported a lower number of pores at the start of sequencing than the number reported by the Flow Cell Check | An air bubble was introduced into the nanopore array | After the Flow Cell Check it is essential to remove any air bubbles near the priming port before priming the flow cell. If not removed, the air bubble can travel to the nanopore array and irreversibly damage the nanopores that have been exposed to air. The best practice to prevent this from happening is demonstrated in videos for how to load a MinION Flow Cell and how to load a PromethION Flow Cell. |
| MinKNOW reported a lower number of pores at the start of sequencing than the number reported by the Flow Cell Check | The flow cell is not correctly inserted into the device | Stop the sequencing run, remove the flow cell from the sequencing device and insert it again, checking that the flow cell is firmly seated in the device and that it has reached the target temperature. If applicable, try a different position on the device (GridION/PromethION). |
| MinKNOW reported a lower number of pores at the start of sequencing than the number reported by the Flow Cell Check | Contaminations in the library damaged or blocked the pores | The pore count during the Flow Cell Check is performed using the QC DNA molecules present in the flow cell storage buffer. At the start of sequencing, the library itself is used to estimate the number of active pores. Because of this, variability of about 10% in the number of pores is expected. A significantly lower pore count reported at the start of sequencing can be due to contaminants in the library that have damaged the membranes or blocked the pores. Alternative DNA/RNA extraction or purification methods may be needed to improve the purity of the input material. The effects of contaminants are shown in the Contaminants Know-how piece. Please try an alternative extraction method that does not result in contaminant carryover. |
MinKNOW script failed
| Observation | Possible cause | Comments and actions |
|---|---|---|
| MinKNOW shows "Script failed" | Restart the computer and then restart MinKNOW. If the issue persists, please collect the MinKNOW log files and contact Technical Support. If you do not have another sequencing device available, we recommend storing the flow cell and the loaded library at 4°C and contact Technical Support for further storage guidance. |
Pore occupancy below 40%
| Observation | Possible cause | Comments and actions |
|---|---|---|
| Pore occupancy <40% | Not enough library was loaded on the flow cell | 10–20 fmol of good quality library can be loaded on to a MinION/GridION flow cell. Please quantify the library before loading and calculate mols using tools like the Promega Biomath Calculator, choosing "dsDNA: µg to pmol" |
| Pore occupancy close to 0 | The Native Barcoding Kit was used, and ethanol was used instead of LFB or SFB at the wash step after sequencing adapter ligation | Ethanol can denature the motor protein on the sequencing adapters. Make sure the LFB or SFB buffer was used after ligation of sequencing adapters. |
| Pore occupancy close to 0 | No tether on the flow cell | Tethers are adding during flow cell priming (FCT tube). Make sure FCT was added to FCF before priming. |
Shorter than expected read length
| Observation | Possible cause | Comments and actions |
|---|---|---|
| Shorter than expected read length | Unwanted fragmentation of DNA sample | Read length reflects input DNA fragment length. Input DNA can be fragmented during extraction and library prep. 1. Please review the Extraction Methods in the Nanopore Community for best practice for extraction. 2. Visualise the input DNA fragment length distribution on an agarose gel before proceeding to the library prep. 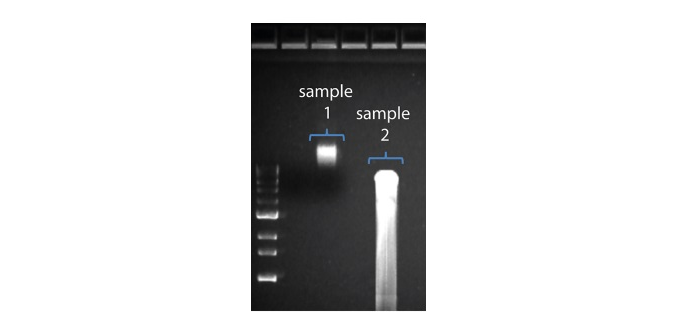 In the image above, Sample 1 is of high molecular weight, whereas Sample 2 has been fragmented. In the image above, Sample 1 is of high molecular weight, whereas Sample 2 has been fragmented.3. During library prep, avoid pipetting and vortexing when mixing reagents. Flicking or inverting the tube is sufficient. |
Large proportion of unavailable pores
| Observation | Possible cause | Comments and actions |
|---|---|---|
Large proportion of unavailable pores (shown as blue in the channels panel and pore activity plot) 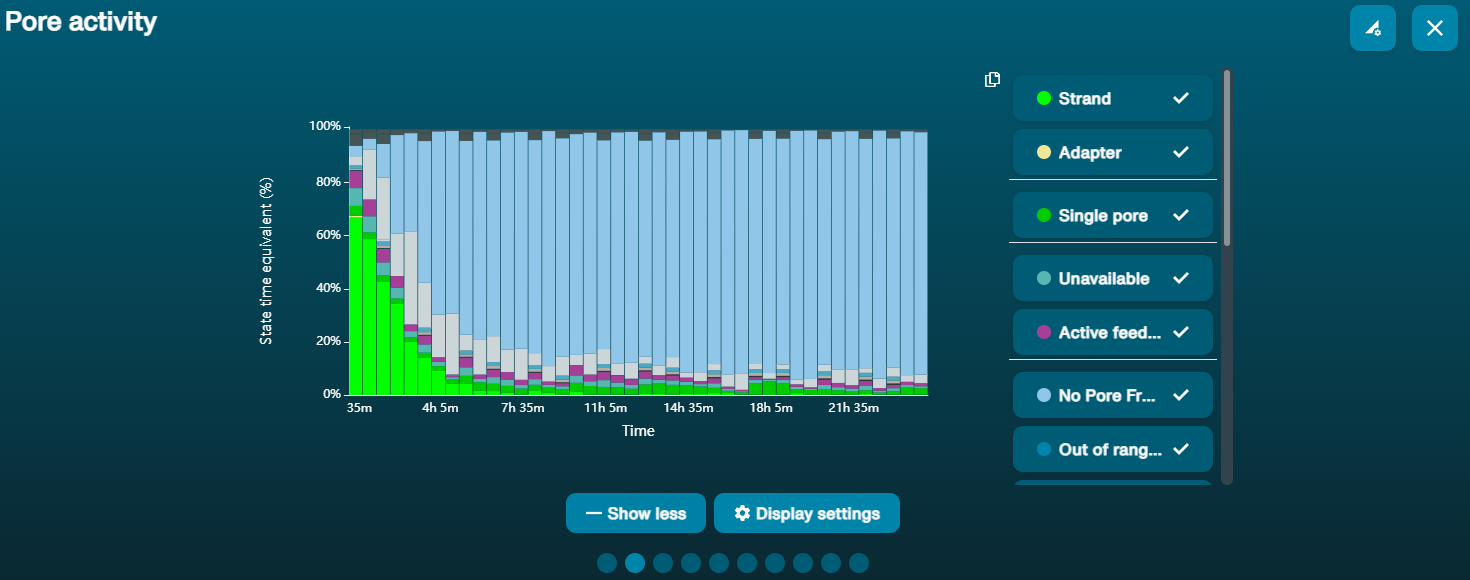 The pore activity plot above shows an increasing proportion of "unavailable" pores over time. The pore activity plot above shows an increasing proportion of "unavailable" pores over time. | Contaminants are present in the sample | Some contaminants can be cleared from the pores by the unblocking function built into MinKNOW. If this is successful, the pore status will change to "sequencing pore". If the portion of unavailable pores stays large or increases: 1. A nuclease flush using the Flow Cell Wash Kit (EXP-WSH004) can be performed, or 2. Run several cycles of PCR to try and dilute any contaminants that may be causing problems. |
Large proportion of inactive pores
| Observation | Possible cause | Comments and actions |
|---|---|---|
| Large proportion of inactive/unavailable pores (shown as light blue in the channels panel and pore activity plot. Pores or membranes are irreversibly damaged) | Air bubbles have been introduced into the flow cell | Air bubbles introduced through flow cell priming and library loading can irreversibly damage the pores. Watch the how to load a MinION Flow Cell or how to load a PromethION Flow Cell videos for best practice. |
| Large proportion of inactive/unavailable pores | Certain compounds co-purified with DNA | Known compounds, include polysaccharides, typically associate with plant genomic DNA. 1. Please refer to the Plant leaf DNA extraction method. 2. Clean-up using the QIAGEN PowerClean Pro kit. 3. Perform a whole genome amplification with the original gDNA sample using the QIAGEN REPLI-g kit. |
| Large proportion of inactive/unavailable pores | Contaminants are present in the sample | The effects of contaminants are shown in the Contaminants Know-how piece. Please try an alternative extraction method that does not result in contaminant carryover. |
Reduction in sequencing speed and q-score later into the run
| Observation | Possible cause | Comments and actions |
|---|---|---|
| Reduction in sequencing speed and q-score later into the run | Fast fuel consumption is typically seen in Kit 9 chemistry (e.g. SQK-LSK109) when the flow cell is overloaded with library. Please see the appropriate protocol for your DNA library to find the recommendation. | Add more fuel to the flow cell by following the instructions in the MinKNOW protocol. In future experiments, load lower amounts of library to the flow cell. |
Temperature fluctuation
| Observation | Possible cause | Comments and actions |
|---|---|---|
| Temperature fluctuation | The flow cell has lost contact with the device | Check that there is a heat pad covering the metal plate on the back of the flow cell. Re-insert the flow cell and press it down to make sure the connector pins are firmly in contact with the device. If the problem persists, please contact Technical Services. |
Failed to reach target temperature
| Observation | Possible cause | Comments and actions |
|---|---|---|
| MinKNOW shows "Failed to reach target temperature" | The instrument was placed in a location that is colder than normal room temperature, or a location with poor ventilation (which leads to the flow cells overheating) | MinKNOW has a default timeframe for the flow cell to reach the target temperature. Once the timeframe is exceeded, an error message will appear and the sequencing experiment will continue. However, sequencing at an incorrect temperature may lead to a decrease in throughput and lower q-scores. Please adjust the location of the sequencing device to ensure that it is placed at room temperature with good ventilation, then re-start the process in MinKNOW. Please refer to this link for more information on MinION temperature control. |







