Cell free DNA (cfDNA) information repository (CFD_S1020_v1_revB_10Mar2025)
InfoSheet
Cell free DNA (cfDNA) information repository V CFD_S1020_v1_revB_10Mar2025
Analysis of cell-free (cf)DNA can be used for a range of uses, including cancer detection and tissue-of-origin analysis.
This document is a repository for all of our internally developed methods, know-how documents and information. This will ensure our users can easily access all the required information and our recommended best practices using nanopore sequencing technology for cfDNA.
FOR RESEARCH USE ONLY
Overview
Analysis of cell-free (cf)DNA can be used for a range of uses, including cancer detection and tissue-of-origin analysis.
This document is a repository for all of our internally developed methods, know-how documents and information. This will ensure our users can easily access all the required information and our recommended best practices using nanopore sequencing technology for cfDNA.
1. Resources
End-to-end protocols
Our end-to-end protocols provide you with a step-by-step guide to carry out the preparation and sequencing of a human cfDNA sample from fresh blood or plasma.
| Ligation sequencing V14 — Human cfDNA singleplex (SQK-LSK114) |
|---|
| Our singleplex method allows you to process a single human cell-free DNA (cfDNA) sample with our Ligation Sequencing Kit V14 (SQK-LSK114). Typically, we obtain ~50 Gb of aligned data (15x coverage) for human cfDNA samples processed with this protocol. |
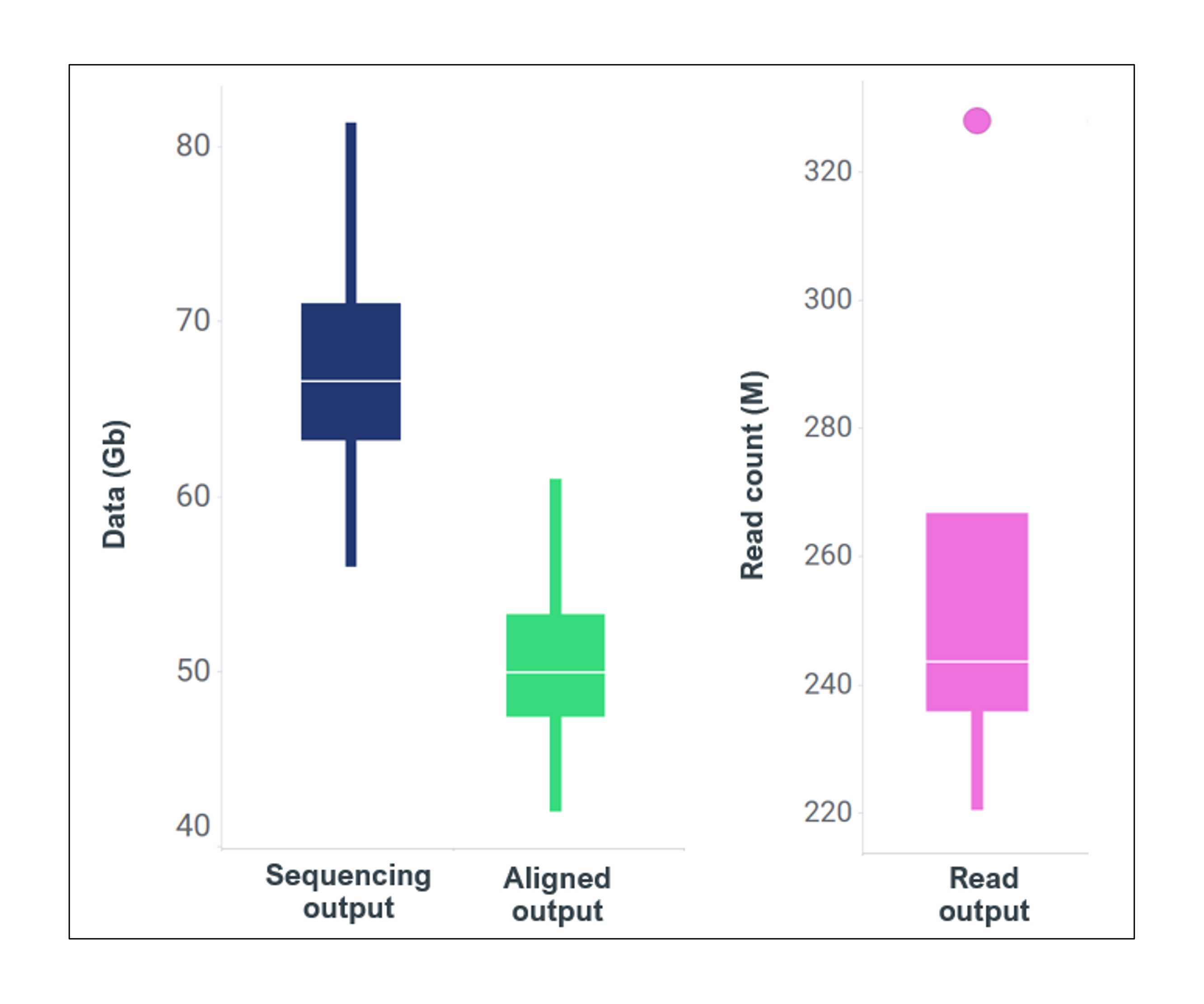 |
| Sequencing and primary aligned output for 4x replicates of ~15 ng inputs, with each library run on a single PromethION flow cell. Where a) sequencing output averages 70 Gb, b) primary aligned data averages 52 Gb and c) read count averages 227 M. |
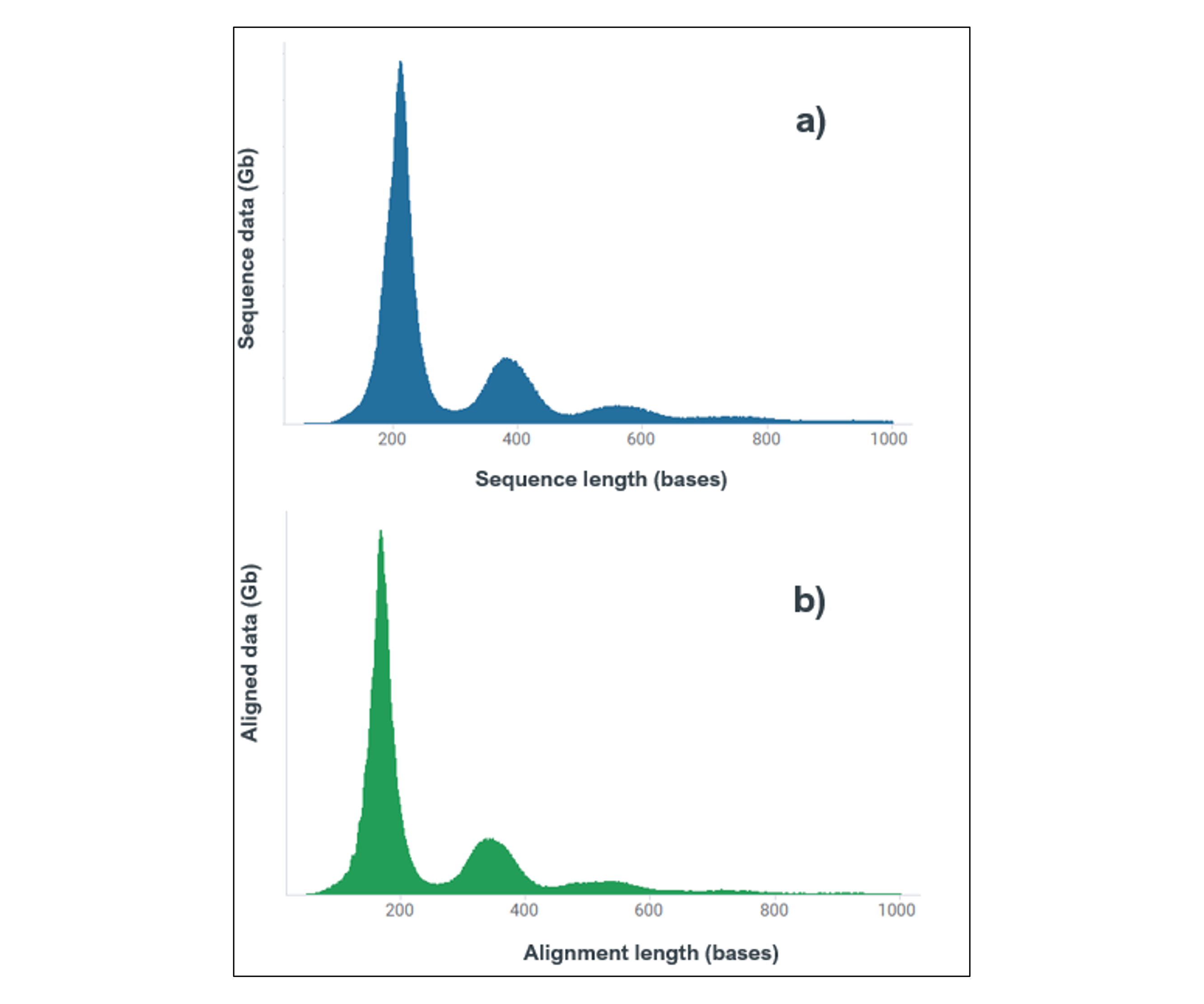 |
| Read length profiles (Q-score ≥10) of total data output for a) Sequencing data, b) Aligned data. The marginal reduction in length of aligned data is due to the informatic removal of sequencing adapter. These profiles are equivalent for each of the 3x replicates. |
| Ligation sequencing V14 — Human cfDNA multiplex (SQK-NBD114.24) |
|---|
| Our multiplex method describes how to carry out preparation and sequencing of 12 human cell-free DNA (cfDNA) samples using the Native Barcoding Kit 24 V14 (SQK-NBD114.24). Typically, we obtain ~3 Gb of aligned data (1x coverage) for each of the 12 human cfDNA samples processed with this protocol. |
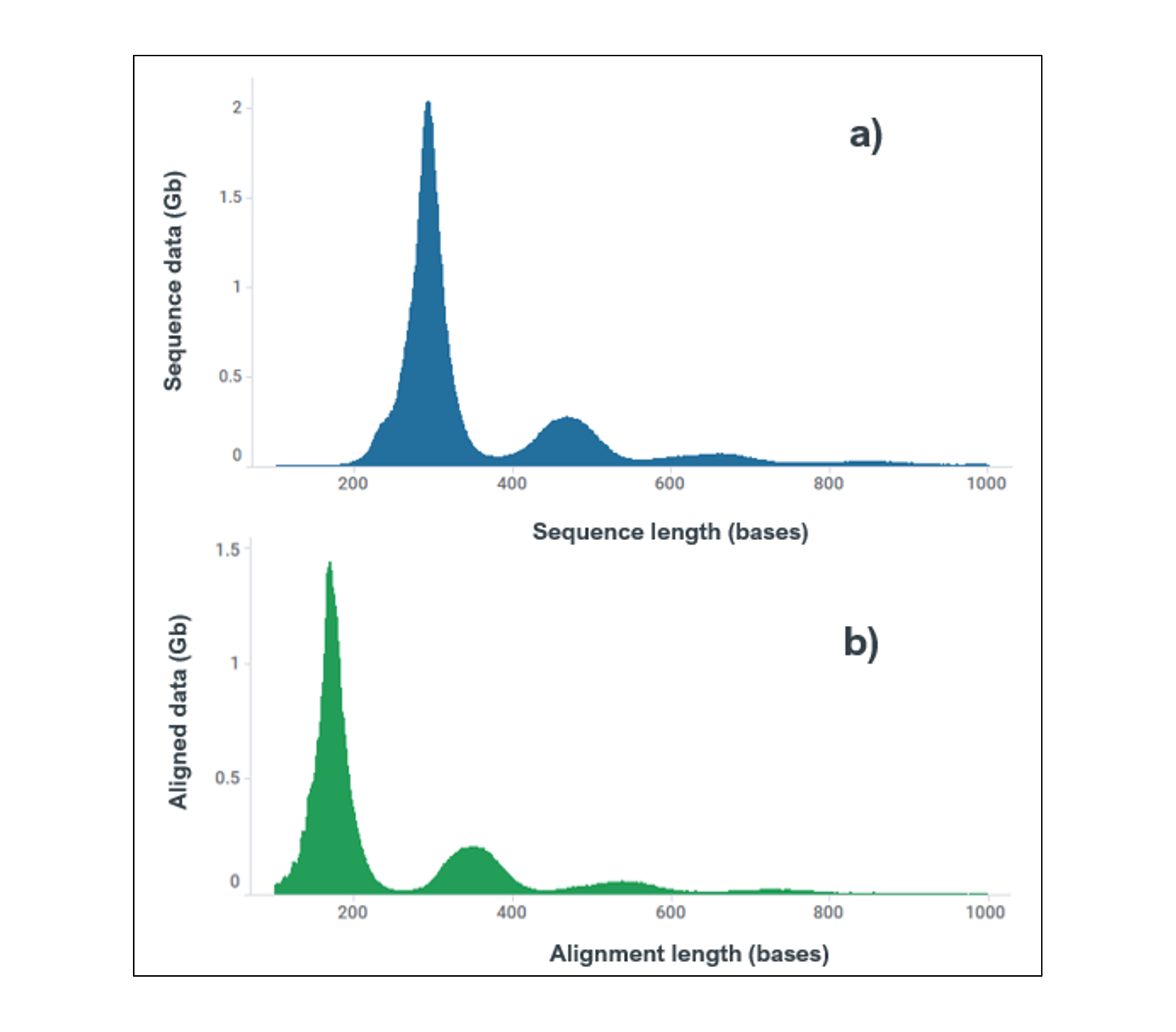 |
| Read length profiles (Q-score ≥10) of total data output (multiplexed) for a) Sequencing data, b) Aligned data. The reduction in length of aligned data is due to the removal of native barcode and adapter sequence. These profiles are equivalent for each of the 12x demultiplexed samples. |
Sample extraction methods
Our extraction methods have been developed by our internal teams to generate the best input available for your sequencing experiment. These methods use the QIAGEN QIAamp MinElute ccfDNA Midi Kit. Please note, these sample extraction methods have also been fully integrated into our end-to-end protocols.
Human blood cell-free DNA (cfDNA) extraction for singleplex sequencing
Human blood cell-free DNA (cfDNA) extraction for multiplex sequencing
 Example of fragment length profile of extracted cfDNA using QIAGEN QIAamp MinElute ccfDNA Midi Kit, run on a Femto Pulse (Agilent). This example shows the characteristic nucleosome peaks with minimal gDNA contamination.
Example of fragment length profile of extracted cfDNA using QIAGEN QIAamp MinElute ccfDNA Midi Kit, run on a Femto Pulse (Agilent). This example shows the characteristic nucleosome peaks with minimal gDNA contamination.
Know-how and information documents
Our know-how and information documents present you with additional background information on the methods we have developed and/or provide additional practices and data to optimise your sequencing experiment.
Updated method for cell-free DNA (cfDNA) methylation profiling:
Document providing additional information and background on the cfDNA sequencing methods for retaining methylation in cfDNA reads, providing bioinformatic options to omit methylation information at specific read positions, and characterising the loss of methylation for the three most common cfDNA nucleosome lengths.Optimisation of library preparation for longer cell-free DNA (cfDNA) libraries:
Study showing that the characteristic length profile of cfDNA can be manipulated by performing size selection during library preparation to enrich for longer fragments. We also show that the fragment length distribution of the sample is a reasonable predictor of both the raw read length and aligned read length profiles observed from sequencing.Assessment of cfDNA extraction kits for nanopore sequencing:
Study assessing the performance of commonly used cfDNA extraction kits with regards to cfDNA yield and sequencing performance.Comparative analysis of cfDNA collection tubes for nanopore sequencing:
Study evaluating the performance of commonly used blood collection tubes and demonstrating how the choice of collection tube and sample storage time can impact sequencing performance.






