Reduced representation methylation multiplex sequencing (RRMS) from cells using SQK-NBD114.24 (RRMS_9209_v114_revI_21Oct2025)
PromethION: Protocol
Reduced representation methylation multiplex sequencing (RRMS) from cells using SQK-NBD114.24 V RRMS_9209_v114_revI_21Oct2025
For Research Use Only.
FOR RESEARCH USE ONLY
Contents
Introduction to the protocol
Sample preparation
Library preparation
- 6. DNA repair and end-prep
- 7. Native barcode ligation
- 8. Adapter ligation and clean-up
- 9. Priming and loading the PromethION Flow Cell
- 10. Washing and reloading a PromethION Flow Cell
Sequencing and data analysis
Troubleshooting
Overview
For Research Use Only.
1. Overview of the protocol
Adaptive sampling in Kit 14 chemistry
While using Kit 14 chemistry, this workflow has been optimised to enrich specific regions of interest (ROIs) with Adaptive sampling, ensuring highest output and the best sequencing results.
For more background information about designing an adaptive sampling experiment, please refer to the Adaptive sampling best practice document: Adaptive sampling best practice
Reduced representation methylation sequencing (RRMS)
Nanopore sequencing enables direct detection of methylated cytosines (e.g., at CpG sites), without the need for bisulphite conversion. CpG sites frequently occur in high density clusters called CpG islands (CGI) and most of vertebrate genes have their promoters embedded within CGIs.
Changes in methylation patterns within promoters is associated with changes in gene expression and disease states such as cancer: exploring methylation differences between tumour samples and normal samples can help to uncover mechanisms associated with tumour formation and development.
Adaptive sampling (AS) offers a fast, flexible and precise method to enrich for regions of interest (e.g. CGIs) by depleting off-target regions during sequencing itself with no requirement for upfront sample manipulation.
To read more about how the method works, and how it compares to other techniques for analysing methylation (e.g. EPIC arrays, bisulfite), please see our Introduction to Reduced-Representation Methylation Sequencing.
RRMS can be deployed on a GridION and PromethION P2S, P24 and P48 devices.
When running on a GridION, we recommend running a single sample per flow cell - using our Reduced representation methylation sequencing (RRMS) from cells using SQK-LSK114 protocol.
Human sample sequencing
The RRMS protocol enables users to target 310 Mb of the human genome which are highly enriched for CpGs including all annotated CpG islands, shores, shelves and >90% of promoter regions (100% of promoter with more than 4 CpGs). As well as other rich CpG regions in the genome. The total number of CpG sites in the BED file is 7.18 million.
For benchmarking purposes, we performed RRMS on five replicates of a metastatic melanoma cell line and its normal pair for a male individual (COLO829/COLO829_BL) and a triple negative breast cancer cell line pair (HCC1395/HCC1935_BL). Each sample was run on a single MinION/GridION Flow Cell. RRMS resulted in high-confidence methylation calls (>10 overlapping reads) for 7.3-8.5 million CpGs per sample.
For comparison, we also performed Reduced Representation Bisulphite Sequencing (RRBS), which typically yields 1.7–2.5 high confidence calls per sample. More information on this comparison can be accessed in our RRMS performance document and poster.
Mouse sample sequencing
The RRMS protocol and a new BED file have also been developed to target 308 Mb of the mouse genome, covering 100% of CpG island and promoter regions; as well as other rich CpG regions in the genome.
The performance of RRMS for mouse samples was characterised on replicates of a blastocyst-derived, embryonic stem cell line (ES-E14TG2a) and a leukemia cell line (BALB/c AMuLV A.3R.1). A non-RRMS library was also run as a control. Each sample was run on a single MinION Flow Cell and RRMS resulted in high-confidence methylation calls (>10X reads per site) for 5.0–5.8 million CpGs per sample in the mouse genome, compared to ~400,000 CpGs in the control library.
Alternative vertebrate genomes could be sequenced using the RRMS protocol and a bespoke BED file.
However, please note Oxford Nanopore Technologies has only validated this method using human and mouse samples.
Introduction to the DNA extraction and multiplex sequencing protocol for RRMS
This protocol describes how to carry out DNA extraction and reduced representation methylation sequencing (RRMS) for up to 4 samples on a single PromethION Flow Cell, using the Native Barcoding Kit (SQK-NBD114.24) and the Adaptive Sampling feature in MinKNOW.
Steps in the sequencing workflow:
Prepare for your experiment
You will need to:
- Ensure you have your sequencing kit, the correct equipment and third-party reagents.
- Download the software for acquiring and analysing your data.
- Ensure that you have the correct BED file for Adaptive Sampling.
- Check your flow cell to ensure it has enough pores for a good sequencing run.
Sample preparation
- Extract your DNA using the QIAGEN Puregene Cell Kit.
- Fragment your DNA using the Covaris g-TUBE, and check its length, quantity and purity. The quality checks performed during the protocol are essential in ensuring experimental success.
Library preparation
The Table below is an overview of the steps required in the library preparation, including timings and optional stopping points.
| Library preparation | Process | Time | Stop option |
|---|---|---|---|
| DNA repair and end-prep | Repair the fragmented DNA and prepare the DNA ends for barcode attachment. | 35 minutes | 4°C overnight |
| Native barcode ligation | Ligate the native barcodes to the DNA ends. | 60 minutes | 4°C overnight |
| Adapter ligation and clean-up | Attach the sequencing adapters to the barcoded DNA ends. | 50 minutes | 4°C short-term storage or for repeated use, such as re-loading your flow cell. -80°C for single-use, long-term storage. We strongly recommend sequencing your library as soon as it is adapted. |
| Priming and loading the flow cell | Prime the flow cell and load the prepared library for sequencing. | 10 minutes | |
| Washing and reloading the flow cell (x2) | Pause your sequencing run. Wash your flow cell with nuclease to remove the previous library load and unblock pores. Prime the flow cell and reload the prepared library to continue sequencing. | 60 minutes (x2) |
Sequencing and analysis
You will need to:
- Start a sequencing run using the MinKNOW software, which will collect raw data from the device and convert it into basecalled reads. While configuring the run, turn on the Adaptive Sampling setting and import a pre-prepared BED file with your regions of interest, along with a FASTA reference file.
- Sequence the sample for a total of 96 hours, with two flow cell washes when the available pore count drops to around 40% of the starting pore count (typically after ~24 hours and the second time after ~48 hours).
- Use Dorado to call modified bases, for more information please refer to the Dorado github page.
- Use the commands recommended at the end of this protocol to aggregate the modified bases and perform CpG island annotation.
Compatibility of this protocol
This protocol should only be used in combination with:
- Native Barcoding Kit 24 V14 (SQK-NBD114.24)
- R10.4.1 PromethION Flow Cells (FLO-PRO114M)
- Flow Cell Wash Kit (EXP-WSH004)
- Sequencing Auxiliary Vials V14 (EXP-AUX003)
- Native Barcoding Expansion V14 (EXP-NBA114)
- PromethION 24/48 device - PromethION IT requirements document
- PromethION 2 Solo device - PromethION 2 Solo IT requirements document
2. Equipment and consumables
Materials
- 5 x 10^6 cells per sample (FOR EXTRACTION)
- 2 µg of fragmented gDNA per sample (FOR LIBRARY PREP)
- Native Barcoding Kit 24 V14 (SQK-NBD114.24)
- Flow Cell Wash Kit (EXP-WSH004)
Consumables
- PromethION Flow Cells
- Puregene Cell Kit (QIAGEN, 158043)
- TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) (Fisher scientific, 10224683)
- 1 x Phosphate-buffered saline (PBS)
- Isopropanol
- g-TUBE™ (Covaris, 520079)
- NEBNext FFPE Repair Mix (NEB, M6630)
- NEBNext Ultra II End repair/dA-tailing Module (NEB, E7546)
- NEB Blunt/TA Ligase Master Mix (NEB, M0367)
- NEBNext® Quick Ligation Module (NEB, E6056)
- Freshly prepared 70% ethanol in nuclease-free water
- Freshly prepared 80% ethanol in nuclease-free water
- Nuclease-free water (e.g. Thermo Scientific, AM9937)
- 15 ml Falcon tubes
- 1.5 ml Eppendorf DNA LoBind tubes
- 0.2 ml thin-walled PCR tubes
- Qubit™ Assay Tubes (Invitrogen, Q32856)
- Qubit dsDNA HS Assay Kit (Invitrogen, Q32851)
- Qubit dsDNA BR Assay Kit (Invitrogen, Q32850)
Equipment
- PromethION device
- PromethION Flow Cell Light Shield
- Centrifuge and rotor suitable for 15 ml Falcon tubes
- Incubator or water bath set at 37°C and 50°C
- Inoculation loop or disposable tweezers for spooling DNA
- Eppendorf 5424 centrifuge (or equivalent)
- Hula mixer (gentle rotator mixer)
- Magnetic separation rack, suitable for 1.5 ml Eppendorf tubes
- Microfuge
- Vortex mixer
- Thermal cycler
- Wide-bore pipette tips
- P1000 pipette and tips
- P200 pipette and tips
- P100 pipette and tips
- P20 pipette and tips
- P10 pipette and tips
- P2 pipette and tips
- Ice bucket with ice
- Timer
Optional equipment
- Agilent Femto Pulse System (or equivalent for read length QC)
- Qubit™ fluorometer (or equivalent for QC check)
The above list of materials, consumables, and equipment is for the extraction method in the sample preparation section, as well as the library preparation section of the protocol. If you have pre-extracted sample(s), you will only require the materials for the library preparation section of this protocol.
For this protocol, the following inputs are required:
Input requirements per sample for the extraction method:
- 5x106 cells per sample
Input requirements per sample for the library preparation:
- 2 µg of g-TUBE fragmented gDNA
Input DNA
How to QC your input DNA
It is important that the input DNA meets the quantity and quality requirements. Using too little or too much DNA, or DNA of poor quality (e.g. highly fragmented or containing RNA or chemical contaminants) can affect your library preparation.
For instructions on how to perform quality control of your DNA sample, please read the Input DNA/RNA QC protocol.
Chemical contaminants
Depending on how the DNA is extracted from the raw sample, certain chemical contaminants may remain in the purified DNA, which can affect library preparation efficiency and sequencing quality. Read more about contaminants on the Contaminants page of the Community.
Third-party reagents
We have validated and recommend the use of all the third-party reagents used in this protocol. Alternatives have not been tested by Oxford Nanopore Technologies.
For all third-party reagents, we recommend following the manufacturer's instructions to prepare the reagents for use.
Check your flow cell
We highly recommend that you check the number of pores in your flow cell prior to starting a sequencing experiment. This should be done within 12 weeks of purchasing your PromethION Flow Cells. Oxford Nanopore Technologies will replace any unused flow cell with fewer than the number of pores listed in the Table below, when the result is reported within two days of performing the flow cell check, and when the storage recommendations have been followed. To do the flow cell check, please follow the instructions in the Flow Cell Check document.
| Flow cell | Minimum number of active pores covered by warranty |
|---|---|
| PromethION Flow Cell | 5,000 |
The Native Adapter (NA) included in this kit and protocol is not interchangeable with other sequencing adapters.
Native Barcoding Kit 24 V14 (SQK-NBD114.24) contents
Note: We are in the process of updating our native barcoding kits with an increased volume of Short Fragment Buffer (SFB). If you have an old format kit and/or require additional volume of Short Fragment Buffer (SFB), this can be purchased via our SFB Expansion (EXP-SFB001).
New format: increased volume of Short Fragment Buffer (SFB)
| Name | Acronym | Cap colour | No. of vials | Fill volume per vial (µl) |
|---|---|---|---|---|
| DNA Control Sample | DCS | Yellow | 2 | 35 |
| Native Adapter | NA | Green | 1 | 40 |
| Sequencing Buffer | SB | Red | 1 | 700 |
| Library Beads | LIB | Pink | 1 | 600 |
| Library Solution | LIS | White cap, pink label | 1 | 600 |
| Elution Buffer | EB | Black | 2 | 500 |
| AMPure XP Beads | AXP | Clear cap, light teal label | 1 | 6,000 |
| Long Fragment Buffer | LFB | Orange | 1 | 1,800 |
| Short Fragment Buffer | SFB | Clear | 1 | 13,000 |
| EDTA | EDTA | Blue | 1 | 700 |
| Flow Cell Flush | FCF | Clear cap, light blue label | 1 | 8,000 |
| Flow Cell Tether | FCT | Purple | 1 | 200 |
| Native Barcode plate | NB01-24 | - | 2 plates, 3 sets of barcodes per plate | 5 µl per well |
Old format: lower volume of Short Fragment Buffer (SFB)
| Name | Acronym | Cap colour | No. of vials | Fill volume per vial (µl) |
|---|---|---|---|---|
| DNA Control Sample | DCS | Yellow | 2 | 35 |
| Native Adapter | NA | Green | 1 | 40 |
| Sequencing Buffer | SB | Red | 1 | 700 |
| Library Beads | LIB | Pink | 1 | 600 |
| Library Solution | LIS | White cap, pink label | 1 | 600 |
| Elution Buffer | EB | Black | 2 | 500 |
| AMPure XP Beads | AXP | Clear cap, light teal label | 1 | 6,000 |
| Long Fragment Buffer | LFB | Orange | 1 | 1,800 |
| Short Fragment Buffer | SFB | Clear | 1 | 1,800 |
| EDTA | EDTA | Blue | 1 | 700 |
| Flow Cell Flush | FCF | Clear cap, light blue label | 1 | 8,000 |
| Flow Cell Tether | FCT | Purple | 1 | 200 |
| Native Barcode plate | NB01-24 | - | 2 plates, 3 sets of barcodes per plate | 5 µl per well |
Note: This product contains AMPure XP reagent manufactured by Beckman Coulter, Inc. and can be stored at -20°C with the kit without detriment to reagent stability.
Note: The DNA Control Sample (DCS) is a 3.6 kb standard amplicon mapping the 3' end of the Lambda genome.
3. BED file
Download the BED file from the Adaptive Sampling catalogue.
The Adaptive Sampling catalogue provides a way for both the Oxford Nanopore team and Community members to share BED files with genomic target regions used for Adaptive Sampling experiments. The BED files along with a reference genome can be uploaded into MinKNOW.
For human genome RRMS experiments, download the Human reduced representation methylation sequencing (RRMS) file.
For mouse genome RRMS experiments, download the Mouse reduced representation methylation sequencing (RRMS) file.
(Optional): For alternative vertebrate genomes, please use a bespoke BED file for the desired organism.
4. DNA extraction
Materials
- 5 x 10^6 cells
Consumables
- Puregene Cell Kit (QIAGEN, 158043)
- Freshly prepared 70% ethanol in nuclease-free water
- TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) (Fisher scientific, 10224683)
- 1 x Phosphate-buffered saline (PBS)
- Isopropanol
- Qubit™ dsDNA HS Assay Kit (ThermoFisher, Q32851)
- Qubit™ Assay Tubes (Invitrogen, Q32856)
- 15 ml Falcon tubes
- 1.5 ml Eppendorf DNA LoBind tubes
Equipment
- Centrifuge and rotor suitable for 15 ml Falcon tubes
- Incubator or water bath set at 37°C and 50°C
- Vortex mixer
- Inoculation loop or disposable tweezers for spooling DNA
- Wide-bore pipette tips
- P1000 pipette and tips
- P200 pipette and tips
- P100 pipette and tips
- P20 pipette and tips
- Qubit™ fluorometer (or equivalent for QC check)
Extraction from cultured cell lines:
Extract DNA from your sample(s) using one of our recommended extraction protocols.
For the benchmarking of this method, the Oxford Nanopore team extracted DNA from ~5 million cells using the protocol: Human cell line DNA – QIAGEN Puregene Cell Kit. The steps for this method are outlined below.
Note: this method is also suitable for mouse cell line DNA.
We also offer multiple mammalian sample extraction protocols, which you can use for other sample types.
Harvest and pellet 5 x 10^6 cells by centrifugation at 300 x g for 3 minutes. If any liquid remains associated with the pellet, spin down the cells again and aspirate the remaining supernatant.
Add 200 µl of 1x PBS to the pelleted cells and centrifuge at 300 x g for 3 minutes. Aspirate and discard the supernatant.
Add 2 ml of Cell Lysis Solution to the washed cell pellet. Using a wide-bore pipette tip, resuspend the cells and transfer them to a 15 ml Falcon tube. If clumps of cells remain, gently invert the tube.
Incubate the sample at 37°C for 30 minutes.
Add 700 µl of the Protein Precipitation Solution to the lysed cells and mix by vortexing for three pulses of 5 seconds.
Centrifuge the sample at 2000 x g for 5 minutes.
Transfer the supernatant to a new tube and add 2.5 ml of room temperature isopropanol. Discard the pellet.
Mix by gently inverting the tube 50 times.
Spool the DNA using an inoculation loop or disposable tweezers.
Dip the spooled DNA in an Eppendorf tube containing 70% cold ethanol.
Remove the inoculation loop or tweezers with the spooled DNA from the ethanol tube, and allow it to air-dry for a few seconds.
Dip the DNA in a 1.5 ml Eppendorf DNA LoBind tube containing 250 µl TE (1 mM EDTA, pH 8.0) and allow the DNA to gently dislodge from the loop/tweezers.
Incubate the tube for 2 hours at 50°C, occasionally pipette mixing the whole volume tube contents (200 μl) with a wide-bore pipette tip.
Note: The DNA pellet may take some time to solubilise. Please ensure the solution is homogenous before quantifying.
Optional: Alternatively, this incubation can be performed at room temperature overnight.
Quantify 1 µl of each eluted sample using a Qubit fluorometer.
Take forward 2 µg of extracted gDNA, for each sample, into the fragmentation of extracted DNA stage of the protocol.
5. DNA fragmentation
Materials
- 2 µg of extracted gDNA (from previous step)
Consumables
- g-TUBE™ (Covaris, 520079)
- TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) (Fisher scientific, 10224683)
- Qubit dsDNA BR Assay Kit (Invitrogen, Q32850)
- Qubit™ Assay Tubes (Invitrogen, Q32856)
- 1.5 ml Eppendorf DNA LoBind tubes
Equipment
- Eppendorf 5424 centrifuge (or equivalent)
- P1000 pipette and tips
- P200 pipette and tips
- P100 pipette and tips
- P20 pipette and tips
- P2 pipette and tips
- Qubit™ fluorometer (or equivalent for QC check)
Optional equipment
- Agilent Femto Pulse System (or equivalent for read length QC)
Fragmentation of extracted DNA using Covaris g-TUBE:
To prepare fragmented gDNA for the library prep protocol, mechanical fragmentation is performed using a g-TUBE (Covaris) to shear DNA to a fragment length of approximately 6kb.
Prepare the DNA in TE buffer:
- Ensure you have 2 µg of extracted gDNA from the sample extraction, and transfer this into a 1.5 ml Eppendorf tube.
- Adjust the volume to 50 μl with TE buffer.
- Mix thoroughly by pipetting up and down.
- Spin down briefly in a microfuge.
Load the 50 µl of the sample into the top of the g-TUBE. Screw the cap firmly and centrifuge at 11,000 rpm (~11,300 RCF) for 30 seconds.
After centrifugation, spin the tube again at 11,000 rpm (~11,300 RCF) for 10 seconds to ensure complete passage of all gDNA through the constriction.
Visually inspect to confirm the entire sample has passed through the upper chamber to the lower chamber of the g-TUBE.
Invert the g-TUBE and spin it again at the same speed and duration as above: 11,000rpm (~11,300 RCF) for 30 seconds.
Repeat the centrifugation at 11,000 rpm (~11,300 RCF) for 10 seconds to ensure thorough passage of all gDNA through the constriction.
Unscrew the tube body, leaving the screw-cap containing the sample. Retrieve the sample from the g-TUBE screw-cap and transfer it into a clean 1.5 ml Eppendof tube.
Quantify 1 µl of the fragmented gDNA using the Qubit dsDNA Broad Range Assay Kit.
Sample concentration after g-TUBE shearing is expected to be ~40 ng/µl.
The fragmented gDNA should also be assessed using Femto-Pulse (Agilent) to evaluate the size and quality of the DNA.

Example DNA fragment distribution after g-TUBE fragmentation, analysed using an Agilent 165 kb Femto-Pulse Assay. Note the single prominent peak ~6 kb.
Take forward 48 µl of fragmented gDNA (containing ~2 µg), for each sample, into the library preparation section of the protcol.
6. DNA repair and end-prep
Materials
- 2 µg of fragmented gDNA in 48 µl per sample
- AMPure XP Beads (AXP)
Consumables
- NEBNext® FFPE DNA Repair Mix (NEB, M6630)
- NEBNext® Ultra™ II End Repair/dA-Tailing Module (NEB, E7546)
- Qubit dsDNA HS Assay Kit (Invitrogen, Q32851)
- Nuclease-free water (e.g. Thermo Scientific, AM9937)
- Freshly prepared 80% ethanol in nuclease-free water
- Qubit™ Assay Tubes (Invitrogen, Q32856)
- 0.2 ml thin-walled PCR tubes
- 1.5 ml Eppendorf DNA LoBind tubes
Equipment
- P1000 pipette and tips
- P100 pipette and tips
- P10 pipette and tips
- Microfuge
- Thermal cycler
- Hula mixer (gentle rotator mixer)
- Magnetic separation rack
- Ice bucket with ice
Optional equipment
- Qubit™ fluorometer (or equivalent for QC check)
Prepare the NEBNext FFPE DNA Repair Mix and NEBNext Ultra II End Repair / dA-tailing Module reagents in accordance with manufacturer’s instructions, and place on ice.
For optimal performance, NEB recommend the following:
Thaw all reagents on ice.
Flick and/or invert the reagent tubes to ensure they are well mixed.
Note: Do not vortex the FFPE DNA Repair Mix or Ultra II End Prep Enzyme Mix.Always spin down tubes before opening for the first time each day.
The Ultra II End Prep Reaction Buffer and FFPE DNA Repair Buffer may have a little precipitate. Allow the mixture to come to room temperature and pipette the buffer up and down several times to break up the precipitate, followed by vortexing the tube for 30 seconds to solubilise any precipitate.
Note: It is important the buffers are mixed well by vortexing.The FFPE DNA Repair Buffer may have a yellow tinge and is fine to use if yellow.
Prepare your DNA samples in nuclease-free water:
For each sample, ensure you have ~2 μg of extracted and fragmented DNA from the sample extraction in separate 1.5 ml Eppendorf DNA LoBind tubes.
If necessary, adjust the volume to 48 μl with nuclease-free water.
Mix thoroughly by pipetting up and down, or by flicking the tube.
Spin down briefly in a microfuge.
In each of the 1.5 ml Eppendorf DNA LoBind tubes containing your fragmented DNA samples, mix the following:
| Reagent | Volume |
|---|---|
| ~2 µg of fragmented DNA from the previous step | 48 µl |
| Ultra II End-prep Reaction Buffer | 3.5 µl |
| Ultra II End-prep Enzyme Mix | 3 µl |
| NEBNext FFPE DNA Repair Buffer | 3.5 µl |
| NEBNext FFPE DNA Repair Mix | 2 µl |
| Total | 60 µl |
Thoroughly mix the reaction by gently pipetting and briefly spinning down.
Using a thermal cycler with a heated lid, incubate the reaction at 20°C for 5 minutes, 65°C for 5 minutes and hold at 4°C.
Remove the reaction from the thermal cycler and place the tube on ice.
Resuspend the AMPure XP Beads (AXP) by vortexing.
Add a 1x volume (60 µl) of resuspended the AMPure XP Beads (AXP) to each end-prep reaction and mix by flicking the tube.
Incubate on a Hula mixer (rotator mixer) for 5 minutes at room temperature.
Prepare 3 ml of fresh 80% ethanol in nuclease-free water.
Spin down the sample and pellet on a magnet until supernatant is clear and colourless. Keep the tube on the magnet, and pipette off and discard the supernatant.
Keep the tube on the magnet and wash the beads with 200 µl of freshly prepared 80% ethanol without disturbing the pellet. Remove the ethanol using a pipette and discard.
If the pellet was disturbed, wait for beads to pellet again before removing the ethanol.
Repeat the previous step.
Spin down and place the tube back on the magnet. Pipette off any residual ethanol. Allow to dry for ~30 seconds, but do not dry the pellet to the point of cracking.
For each sample, remove the tube from the magnetic rack and resuspend the pellet in 20 µl nuclease-free water. Incubate for 2 minutes at room temperature.
Pellet the beads on a magnet until the eluate is clear and colourless, for at least 1 minute.
For each sample, remove and retain 20 µl of eluate into a separate clean 1.5 ml Eppendorf DNA LoBind tube.
Note: Ensure your samples are processed separately. At this stage they are not yet barcoded.
Quantify each eluted sample using a Qubit fluorometer.
Note: We recommend performing multiple (triplicate) Qubit readings of each sample to quantify them more accurately. This will be essential for further normalising of each sample before barcoding.
You should expect to recover 1000–1600 ng per sample after end-prep.
Using your quantification results, normalise your samples to the sample with the lowest yield.
- Take forward 15 µl of your lowest performing sample into a clean 0.2 ml thin-walled PCR tube.
- Take forward an equivalent mass of each of the other samples into separate clean 0.2 ml thin-walled PCR tubes.
- Adjust the volume of each of the samples 15 µl using nuclease-free water.
Take forward the equimolar mass of repaired and end-prepped DNA samples in 15 µl into the native barcode ligation step. However, at this point it is also possible to store the sample at 4°C overnight.
7. Native barcode ligation
Materials
- End-prepped DNA in 15 µl from previous step
- Native Barcodes (NB01-24)
- AMPure XP Beads (AXP)
- EDTA (EDTA)
- S Fragment Buffer (SFB)
Consumables
- NEB Blunt/TA Ligase Master Mix (NEB, M0367)
- Freshly prepared 80% ethanol in nuclease-free water
- Nuclease-free water (e.g. Thermo Scientific, AM9937)
- 1.5 ml Eppendorf DNA LoBind tubes
- Eppendorf twin.tec® PCR plate 96 LoBind, semi-skirted (Merck, EP0030129504) with heat seals
- OR 0.2 ml thin-walled PCR tubes
- Qubit™ Assay Tubes (Invitrogen, Q32856)
- Qubit™ dsDNA HS Assay Kit (ThermoFisher, Q32851)
Equipment
- Magnetic separation rack
- Vortex mixer
- Hula mixer (gentle rotator mixer)
- Microfuge
- Thermal cycler
- Ice bucket with ice
- Multichannel pipette and tips
- P1000 pipette and tips
- P200 pipette and tips
- P100 pipette and tips
- P20 pipette and tips
- P10 pipette and tips
- P2 pipette and tips
- Qubit™ fluorometer (or equivalent for QC check)
Prepare the NEB Blunt/TA Ligase Master Mix according to the manufacturer's instructions, and place on ice:
Thaw the reagents at room temperature.
Spin down the reagent tubes for 5 seconds.
Ensure the reagents are fully mixed by performing 10 full volume pipette mixes.
Thaw the EDTA at room temperature and mix by vortexing. Then spin down and place on ice.
Thaw the Native Barcodes (NB01-24) at room temperature. Briefly spin down, individually mix the barcodes required for your number of samples by pipetting, and place them on ice.
The wells of the barcoding plate are intended for single use only. Please ensure your barcode well is sealed before use, and do not reuse the barcode well once pierced/opened.
Select a unique barcode for each sample to be run together on the same flow cell. The samples should be barcoded and combined in one experiment.
Please note: Only use one barcode per sample.
In the 0.2 ml PCR-tubes containing your normalised sample inputs, add the reagents in the following order for each sample:
| Reagent | Volume |
|---|---|
| End-prepped DNA | 15 µl |
| Native Barcode (NB01-24) | 5 µl |
| Blunt/TA Ligase Master Mix | 20 µl |
| Total | 40 µl |
Thoroughly mix the reaction by gently pipetting and briefly spinning down.
Incubate for 20 minutes at room temperature.
Add 4 µl EDTA (blue cap) to each tube and mix thoroughly by pipetting and spin down briefly.
EDTA is added at this step to stop the reaction.
Pool all the barcoded samples in a 1.5 ml Eppendorf DNA LoBind tube.
| . | For 4 samples |
|---|---|
| Total volume for preps using EDTA (blue cap) | 176 µl |
We recommend checking the base of your tubes/plate are all the same volume before pooling and after to ensure all the liquid has been taken forward.
Resuspend the AMPure XP Beads (AXP) by vortexing.
Add 0.65X AMPure XP Beads (AXP) to the pooled reaction, and mix by pipetting.
| . | For 4 samples |
|---|---|
| Volume of AXP for preps using EDTA (blue cap) | 115 µl |
Incubate on a Hula mixer (rotator mixer) for 10 minutes at room temperature.
The next clean-up step uses Short Fragment Buffer (SFB) rather than 80% ethanol to wash the beads. The use of ethanol will be detrimental to the sequencing reaction.
Spin down the sample and pellet on a magnet for 5 minutes. Keep the tube on the magnetic rack until the eluate is clear and colourless, and pipette off the supernatant.
Wash the beads by adding 500 μl Short Fragment Buffer (SFB). Flick the beads to resuspend, spin down, then return the tube to the magnetic rack and allow the beads to pellet. Remove the supernatant using a pipette and discard.
Repeat the previous step.
Spin down and place the tube back on the magnetic rack. Pipette off any residual Short Fragment Buffer (SFB). Allow the pellet to dry for ~30 seconds, but do not dry the pellet to the point of cracking.
Remove the tube from the magnetic rack and resuspend the pellet in 32 µl nuclease-free water by gently flicking.
Incubate for 15 minutes at 37°C. Every 2 minutes, agitate the sample by gently flicking for 10 seconds to encourage DNA elution.
Pellet the beads on a magnetic rack until the eluate is clear and colourless.
Remove and retain 32 µl of eluate into a clean 1.5 ml Eppendorf DNA LoBind tube.
Quantify 1 µl of eluted sample using a Qubit fluorometer.
Note: You should expect to recover between 2200–3200 ng following barcode ligation.
Take forward the barcoded DNA library to the adapter ligation and clean-up step. However, you may store the sample at 4°C overnight.
8. Adapter ligation and clean-up
Materials
- Long Fragment Buffer (LFB)
- Elution Buffer (EB)
- Native Adapter (NA)
- AMPure XP Beads (AXP)
Consumables
- NEBNext® Quick Ligation Module (NEB, E6056)
- 1.5 ml Eppendorf DNA LoBind tubes
- Qubit™ Assay Tubes (Invitrogen, Q32856)
- Qubit™ dsDNA HS Assay Kit (ThermoFisher, Q32851)
Equipment
- Microfuge
- Magnetic separation rack
- Vortex mixer
- Hula mixer (gentle rotator mixer)
- Thermal cycler
- P1000 pipette and tips
- P200 pipette and tips
- P100 pipette and tips
- P20 pipette and tips
- P10 pipette and tips
- Ice bucket with ice
- Qubit™ fluorometer (or equivalent for QC check)
The Native Adapter (NA) used in this kit and protocol is not interchangeable with other sequencing adapters.
Check your flow cell.
We recommend performing a flow cell check before starting adapter ligation and clean-up to ensure you have a flow cell with sufficient pores for a good sequencing run.
See the flow cell check document for more information.
Prepare the NEBNext Quick Ligation Reaction Module according to the manufacturer's instructions, and place on ice:
Thaw the reagents at room temperature.
Spin down the reagent tubes for 5 seconds.
Ensure the reagents are fully mixed by performing 10 full volume pipette mixes. Note: Do NOT vortex the Quick T4 DNA Ligase.
The NEBNext Quick Ligation Reaction Buffer (5x) may have a little precipitate. Allow the mixture to come to room temperature and pipette the buffer up and down several times to break up the precipitate, followed by vortexing the tube for several seconds to ensure the reagent is thoroughly mixed.
Do not vortex the Quick T4 DNA Ligase.
Spin down the Native Adapter (NA) and Quick T4 DNA Ligase, pipette mix and place on ice.
Thaw the Elution Buffer (EB) at room temperature and mix by vortexing. Then spin down and place on ice.
Thaw the Long Fragment Buffer (LFB) at room temperature and mix by vortexing. Then spin down and place on ice.
In a 1.5 ml Eppendorf LoBind tube, mix in the following order:
Between each addition, pipette mix 10 - 20 times.
| Reagent | Volume |
|---|---|
| Pooled barcoded sample | 30 µl |
| Native Adapter (NA) | 5 µl |
| NEBNext Quick Ligation Reaction Buffer (5X) | 10 µl |
| Quick T4 DNA Ligase | 5 µl |
| Total | 50 µl |
Thoroughly mix the reaction by gently pipetting and briefly spinning down.
Incubate the reaction for 20 minutes at room temperature.
The next clean-up step uses Long Fragment Buffer (LFB) rather than 80% ethanol to wash the beads. The use of ethanol will be detrimental to the sequencing reaction.
Resuspend the AMPure XP Beads (AXP) by vortexing.
Add 25 µl (0.5x) of resuspended AMPure XP Beads (AXP) to the reaction and mix by pipetting.
Incubate on a Hula mixer (rotator mixer) for 10 minutes at room temperature.
Spin down the sample and pellet on the magnetic rack. Keep the tube on the magnet and pipette off the supernatant.
Wash the beads by adding 250 μl Long Fragment Buffer (LFB). Flick the beads to resuspend, spin down, then return the tube to the magnetic rack and allow the beads to pellet for at least 5 minutes. Remove the supernatant using a pipette and discard.
Note: Take care when removing the supernatant, the viscosity of the buffer can contribute to loss of beads from the pellet.
Repeat the previous step.
Spin down and place the tube back on the magnet. Pipette off any residual supernatant. Allow to dry for ~30 seconds, but do not dry the pellet to the point of cracking.
Remove the tube from the magnetic rack and resuspend the pellet in 97 µl of Elution Buffer (EB).
Spin down and incubate for 20 minutes at 37°C. Every 2 minutes, agitate the sample by gently flicking for 10 seconds to encourage DNA elution.
Pellet the beads on a magnet until the eluate is clear and colourless, for at least 1 minute.
Remove and retain 97 µl of eluate containing the DNA library into a clean 1.5 ml Eppendorf DNA LoBind tube.
Dispose of the pelleted beads
Quantify 1 µl of eluted sample using a Qubit fluorometer.
Note: You should expect to recover 1000–1200 ng after adapter ligation and clean-up in a volume of 96 µl.
The prepared library is used for loading into the flow cell. Store the library on ice or at 4°C until ready to load.
Library storage recommendations
We recommend storing libraries in Eppendorf DNA LoBind tubes at 4°C for short-term storage or repeated use, for example, re-loading flow cells between washes. For single use and long-term storage of more than 3 months, we recommend storing libraries at -80°C in Eppendorf DNA LoBind tubes.
9. Priming and loading the PromethION Flow Cell
Materials
- Sequencing Buffer (SB)
- Library Beads (LIB)
- Flow Cell Tether (FCT)
- Flow Cell Flush (FCF)
Consumables
- PromethION Flow Cells
- 1.5 ml Eppendorf DNA LoBind tubes
Equipment
- PromethION 2 Solo or PromethION 24/48 device
- PromethION device
- PromethION Flow Cell Light Shield
- P1000 pipette and tips
- P200 pipette and tips
- P20 pipette and tips
This kit is only compatible with R10.4.1 flow cells (FLO-PRO114M).
After taking the flow cell out of the fridge, wait 20 minutes for the flow cell to reach room temperature before inserting it into the PromethION. Condensation can form on the flow cell in humid environments. Inspect the gold connector pins on the top and underside of the flow cell for condensation and wipe off with a lint-free wipe if any is observed. Ensure the heat pad (black pad) is present on the underside of the flow cell.
Thaw the Sequencing Buffer (SB), Library Beads (LIB), Flow Cell Tether (FCT) and Flow Cell Flush (FCF) at room temperature before mixing by vortexing. Then spin down and store on ice.
To prepare the flow cell priming mix, combine Flow Cell Tether (FCT) and Flow Cell Flush (FCF), as directed below. Mix by vortexing at room temperature.
In a clean suitable tube for the number of flow cells, combine the following reagents:
| Reagent | Volume per flow cell |
|---|---|
| Flow Cell Flush (FCF) | 1,170 µl |
| Flow Cell Tether (FCT) | 30 µl |
| Total volume | 1,200 µl |
For PromethION 2 Solo, load the flow cell(s) as follows:
Place the flow cell flat on the metal plate.
Slide the flow cell into the docking port until the gold pins or green board cannot be seen.

For the PromethION 24/48, load the flow cell(s) into the docking ports:
- Line up the flow cell with the connector horizontally and vertically before smoothly inserting into position.
- Press down firmly onto the flow cell and ensure the latch engages and clicks into place.


Insertion of the flow cells at the wrong angle can cause damage to the pins on the PromethION and affect your sequencing results. If you find the pins on a PromethION position are damaged, please contact support@nanoporetech.com for assistance.

Complete a flow cell check to assess the number of pores available before loading the library.
This step can be omitted if the flow cell has been checked previously.
See the flow cell check document for more information.
Slide the inlet port cover clockwise to open.

Take care when drawing back buffer from the flow cell. Do not remove more than 20-30 µl, and make sure that the array of pores are covered by buffer at all times. Introducing air bubbles into the array can irreversibly damage pores.
After opening the inlet port, draw back a small volume to remove any air bubbles:
- Set a P1000 pipette tip to 200 µl.
- Insert the tip into the inlet port.
- Turn the wheel until the dial shows 220-230 µl, or until you see a small volume of buffer entering the pipette tip.

Load 500 µl of the priming mix into the flow cell via the inlet port, avoiding the introduction of air bubbles. Wait five minutes. During this time, prepare the library for loading using the next steps in the protocol.

Thoroughly mix the contents of the Library Beads (LIB) by pipetting.
The Library Beads (LIB) tube contains a suspension of beads. These beads settle very quickly. It is vital that they are mixed immediately before use.
We recommend using the Library Beads (LIB) for most sequencing experiments. However, the Library Solution (LIS) is available for more viscous libraries.
In a new 1.5 ml Eppendorf DNA LoBind tube, prepare the library for loading as follows:
| Reagent | Volume per flow cell |
|---|---|
| Sequencing Buffer (SB) | 100 µl |
| Library Beads (LIB) thoroughly mixed before use | 68 µl |
| DNA library | 32 µl |
| Total | 200 µl |
Note: The prepared library is used for loading into the flow cell. Store the library on ice or at 4°C until ready to load.
Complete the flow cell priming by slowly loading 500 µl of the priming mix into the inlet port.

Mix the prepared library gently by pipetting up and down just prior to loading.
Load 200 µl of library into the inlet port using a P1000 pipette.

Close the valve to seal the inlet port.
For optimal sequencing output, install the light shield on your flow cell as soon as the library has been loaded.
We recommend leaving the light shield on the flow cell when library is loaded, including during any washing and reloading steps. The shield can be removed when the library has been removed from the flow cell.
If the light shield has been removed from the flow cell, install the light shield as follows:
- Align the inlet port cut out of the light shield with the inlet port cover on the flow cell. The leading edge of the light shield should sit above the flow cell ID.
- Firmly press the light shield around the inlet port cover. The inlet port clip will click into place underneath the inlet port cover.


Close the PromethION lid when ready to start a sequencing run on MinKNOW.
Wait a minimum of 10 minutes after loading the flow cells onto the PromethION before initiating any experiments. This will help to increase the sequencing output.
For instructions on setting up your sequencing run please visit the Data acquisition and basecalling section of this protocol.
Reminder: For this protocol, we recommend washing and reloading your flow cell with fresh library to maintain high data acquisition after ~24 hours of sequencing.
Follow the instructions in the Washing and reloading a PromethION Flow Cell section of this protocol.
10. Washing and reloading a PromethION Flow Cell
Materials
- Adapter ligated DNA library (from previous section)
- Flow Cell Wash Kit (EXP-WSH004)
- Sequencing Auxiliary Vials V14 (EXP-AUX003)
Consumables
- 1.5 ml Eppendorf DNA LoBind tubes
Equipment
- P1000 pipette and tips
- P20 pipette and tips
- Ice bucket with ice
- Vortex mixer
We recommend washing and reloading the flow cell after ~24 hours of sequencing.
For this method, the flow cell is washed after ~24 hours of sequencing to restore pores to ensure efficient data acquisition. After an additional 24 hours of sequencing, the flow cell is washed and reloaded a second time. For this reason, enough library was generated for 3 flow cell loads in the adapter ligation step of the protocol.
- This washing procedure aims to remove most of the initial library and unblock the pores to prepare the flow cell for the loading of a subsequent library.
- Data acquisition in MinKNOW should be paused during the wash procedure and library loading.
- After the flow cell has been washed, the next library can be loaded.
You can navigate to the Pore Activity or the Pore Scan Results plot to see pore availability.
Below you can find example data for pore states observed on a flow cell before and after wash steps are performed. Additionally, you can observe an example for the cummulative sequencing data output, including the wash and reload steps. The red asterisks indicate the flow cell wash and reloads.

Figure 1. Channel state for a 96 hour run. The flow cell washes are incorporated into the method to restore blocked pores, to allow continuous data acquisition. Red asterisks denote when a flush was performed.

Figure 2. Cumulative sequencing data output, over a 96 hour run. Red asterisks denote when a flush was performed.
Washing and reloading a PromethION Flow Cell video
This video will show you how to wash a flow cell after a sequencing run and how to load a new library.
We recommend keeping the light shield on the flow cell during washing if a second library will be loaded straight away.
If the flow cell is to be washed and stored, the light shield can be removed.
Place the tube of Wash Mix (WMX) on ice. Do not vortex the tube.
Thaw one tube of Wash Diluent (DIL) at room temperature.
Mix the contents of Wash Diluent (DIL) thoroughly by vortexing, then spin down briefly and place on ice.
In a fresh 1.5 ml Eppendorf DNA LoBind tube, prepare the following Flow Cell Wash Mix:
| Reagent | Volume per flow cell |
|---|---|
| Wash Mix (WMX) | 2 μl |
| Wash Diluent (DIL) | 398 μl |
| Total | 400 μl |
Mix well by pipetting, and place on ice. Do not vortex the tube.
Pause the sequencing experiment in MinKNOW, and leave the flow cell in the device.
It is vital that the inlet port is closed before removing waste to prevent air from being drawn across the sensor array area, which would lead to a significant loss of sequencing channels.
Remove waste buffer, as follows:
- Close the inlet port.
- Insert a P1000 pipette into a waste port and remove the waste buffer.
Note: As both the inlet port is closed, no fluid should leave the sensor array area.
Slide the inlet port cover clockwise to open the inlet port.

Take care when drawing back buffer from the flow cell. Do not remove more than 20-30 µl, and make sure that the array of pores are covered by buffer at all times. Introducing air bubbles into the array can irreversibly damage pores.
After opening the inlet port, check for a small air bubble under the cover. Draw back a small volume to remove any bubbles:
- Set a P1000 pipette to 200 µl
- Insert the tip into the inlet port
- Turn the wheel until the dial shows 220-230 µl, or until you can see a small volume of buffer entering the pipette tip.

Slowly load 200 µl of the prepared flow cell wash mix into the inlet port, as follows:
- Using a P1000 pipette, take 200 µl of the flow cell wash mix
- Insert the pipette tip into the inlet port, ensuring there are no bubbles in the tip
- Slowly twist the pipette wheel down to load the flow cell (if possible with your pipette) or push down the plunger very slowly, leaving a small volume of buffer in the pipette tip.
- Set a timer for a 5 minute incubation.
Once the 5 minute incubation time is complete, carefully load the remaining 200 µl of the prepared flow cell wash mix into the inlet port, as follows:
- Using a P1000 pipette, take 200 µl of the flow cell wash mix
- Insert the pipette tip into the inlet port, ensuring there are no bubbles in the tip
- Slowly twist the pipette wheel down to load the flow cell (if possible with your pipette) or push down the plunger very slowly, leaving a small volume of buffer in the pipette tip.
Close the inlet port and wait for 1 hour.
It is vital that the inlet port is closed before removing waste to prevent air from being drawn across the sensor array area, which would lead to a significant loss of sequencing channels.
Remove the waste buffer, as follows:
- Ensure the inlet port is closed.
- Insert a P1000 pipette into a waste port and remove the waste buffer
Note: As the inlet port is closed, no fluid should leave the sensor array area.
The buffers used in this process are incompatible with conducting a Flow Cell Check step prior to loading the subsequent library. However, number of available pores will be reported after the next pore scan.
Thaw the Sequencing Buffer (SB), Library Beads (LIB) or Library Solution (LIS, if using), Flow Cell Tether (FCT) and Flow Cell Flush (FCF) at room temperature, before mixing by vortexing. Then spin down before storing on ice.
Prepare the flow cell priming mix in a suitable tube for the number of flow cells to flush. Once combined, mix well by briefly vortexing.
| Reagents | Volume per flow cell |
|---|---|
| Flow Cell Flush (FCF) | 1,170 µl |
| Flow Cell Tether (FCT) | 30 µl |
| Total volume | 1,200 µl |
Slide the inlet port cover clockwise to open.

Take care when drawing back buffer from the flow cell. Do not remove more than 20-30 µl, and make sure that the array of pores are covered by buffer at all times. Introducing air bubbles into the array can irreversibly damage pores.
After opening the inlet port, draw back a small volume to remove any air bubbles:
- Set a P1000 pipette tip to 200 µl.
- Insert the tip into the inlet port.
- Turn the wheel until the dial shows 220-230 µl, or until you see a small volume of buffer entering the pipette tip.

Slowly load 500 µl of the priming mix into the inlet port, as follows:
- Using a P1000 pipette, take 500 µl of the priming mix
- Insert the pipette tip into the priming port, ensuring there are no bubbles in the tip
- Slowly twist the pipette wheel down to load the flow cell (if possible with your pipette) or push down the plunger very slowly, leaving a small volume of buffer in the pipette tip.

It is vital to wait five minutes between the priming mix flushes to ensure effective removal of the nuclease.
Close the inlet port and wait five minutes.
During this time, prepare the library for loading using the next steps in the protocol.
Thoroughly mix the contents of the Library Beads (LIB) by pipetting.
The Library Beads (LIB) tube contains a suspension of beads. These beads settle very quickly. It is vital that they are mixed immediately before use.
We recommend using the Library Beads (LIB) for most sequencing experiments. However, the Library Solution (LIS) is available for more viscous libraries.
In a new 1.5 ml Eppendorf DNA LoBind tube, prepare the library for loading as follows:
| Reagent | Volume per flow cell |
|---|---|
| Sequencing Buffer (SB) | 100 µl |
| Library Beads (LIB) thoroughly mixed before use, or Library Solution (LIS) | 68 µl |
| DNA library | 32 µl |
| Total | 200 µl |
Note: Library loading volume has been increased to improve array coverage.
It is vital that the inlet port is closed before removing waste to prevent air from being drawn across the sensor array area, which would lead to a significant loss of sequencing channels.
Remove the waste buffer, as follows:
- Ensure the inlet port is closed.
- Insert a P1000 pipette into a waste port and remove the waste buffer
Note: As the inlet port is closed, no fluid should leave the sensor array area.
Slide the inlet port cover clockwise to open.

Take care when drawing back buffer from the flow cell. Do not remove more than 20-30 µl, and make sure that the array of pores are covered by buffer at all times. Introducing air bubbles into the array can irreversibly damage pores.
After opening the inlet port, draw back a small volume to remove any air bubbles:
- Set a P1000 pipette tip to 200 µl.
- Insert the tip into the inlet port.
- Turn the wheel until the dial shows 220-230 µl, or until you see a small volume of buffer entering the pipette tip.

Slowly load 500 µl of the priming mix into the inlet port, as follows:
- Using a P1000 pipette, take 500 µl of the priming mix
- Insert the pipette tip into the priming port, ensuring there are no bubbles in the tip
- Slowly twist the pipette wheel down to load the flow cell (if possible with your pipette) or push down the plunger very slowly, leaving a small volume of buffer in the pipette tip.

It is vital that the inlet port is closed before removing waste to prevent air from being drawn across the sensor array area, which would lead to a significant loss of sequencing channels.
Remove waste buffer, as follows:
- Close the inlet port.
- Insert a P1000 pipette into a waste port and remove the waste buffer.
Note: As both the inlet port is closed, no fluid should leave the sensor array area.
Slide the inlet port cover clockwise to open.

Take care when drawing back buffer from the flow cell. Do not remove more than 20-30 µl, and make sure that the array of pores are covered by buffer at all times. Introducing air bubbles into the array can irreversibly damage pores.
After opening the inlet port, draw back a small volume to remove any air bubbles:
- Set a P1000 pipette tip to 200 µl.
- Insert the tip into the inlet port.
- Turn the wheel until the dial shows 220-230 µl, or until you see a small volume of buffer entering the pipette tip.

Mix the prepared library gently by pipetting up and down just prior to loading.
Load 200 µl of library into the inlet port using a P1000 pipette.

Close the valve to seal the inlet port.
For optimal sequencing output, install the light shield on your flow cell as soon as the library has been loaded.
We recommend leaving the light shield on the flow cell when library is loaded, including during any washing and reloading steps. The shield can be removed when the library has been removed from the flow cell.
If the light shield has been removed from the flow cell, install the light shield as follows:
- Align the inlet port cut out of the light shield with the inlet port cover on the flow cell. The leading edge of the light shield should sit above the flow cell ID.
- Firmly press the light shield around the inlet port cover. The inlet port clip will click into place underneath the inlet port cover.


Close the PromethION lid when ready to start a sequencing run on MinKNOW.
Wait a minimum of 10 minutes after loading the flow cells onto the PromethION before initiating any experiments. This will help to increase the sequencing output.
Perform the "Washing and reloading a PromethION flow cell" step twice for a total of three library loads (initial library load + two wash and reloads) to maximise data acquisition.
- The first wash and reload should be performed at ~24 hours into sequencing.
- The second wash and reload should be performed at ~48 hours into sequencing.
11. Data acquisition and basecalling
Overview of nanopore data analysis
For a full overview of nanopore data analysis, which includes options for basecalling and post-basecalling analysis, please refer to the Data Analysis document.
How to start sequencing
The sequencing device control and data acquisition are carried out by the MinKNOW software. Please ensure MinKNOW is installed on your computer. Further instructions for setting up your sequencing run can be found in the MinKNOW protocol.
Sequencing settings for the reduced representation methylation multiplex sequencing protocol:
Select the Native Barcoding Sequencing Kit 24 (SQK-NBD114.24) in kit selection.
Turn basecalling OFF (this will automatically turn off barcoding).
Note: Basecalling and barcoding will be carried out post-sequencing in the downstream analysis section of the protocol.Turn Adaptive Sampling ON, and select Enrich.
Input the human reference file for alignment and the .bed file for enumerating regions (check online catalogue for the human RRMS .bed file).Set the run duration for a minimum of 96 hours.
Set up your desired output parameters.
To ensure the downstream analysis functions correctly, we recommend keeping the default options of the output file format (.POD5).Click Start begin the sequencing run.
12. Downstream analysis
Software versions
See below the software versions used in this guide. Please note, newer versions of the software may not be compatible with commands shown in this guide.
| Software | Version |
|---|---|
| dorado | v0.7.3 |
| modkit | v0.2.8 |
| wf-human-variation | v2.3.0 |
| mosdepth | v0.3.8 |
Basecalling and demux
Basecalling
Dorado stand-alone is used for basecalling using the Dorado basecaller. Open a terminal and enter the following commands:
dorado basecaller hac,5mCG_5hmCG --kit-name SQK-NBD114-24 \
--secondary “no” -Y \
--reference {reference_fasta} {input_pod5_folder} \
| samtools view -e '[qs] >= {qscore_filter}' \
--output {out_pass_bam} \
--unoutput {out_fail_bam}
Notes:
- We recommend using the high accuracy model (hac) for RRMS sequencing runs. However, if using the super accurate model (sup), ensure you are utilizing the correct model in the above command.
- Alignment can be performed while basecalling by providing a reference FASTA file. The recommended human reference file can be downloaded.
- Secondary alignments are discarded by using “--secondary no” and -Y option is enabled, to allow soft-clipping supplementary alignments.
- By providing the option “--kit-name SQK-NBD114-24” Dorado will also classify reads into the different barcodes present by adding a tag to the generated BAM file.
- We recommend setting the qscore filter to 10.
- Please note, GPU compute is needed to perform basecalling with Dorado. More information on how to run Dorado can be found in the github repository.
Demux
Dorado demux is used to sort reads per barcode using the following command:
dorado demux --no-classify --sort-bam --output-dir <out_folder> {out_pass_bam}
Notes:
- This step will generate sorted BAM files for each of the possible barcodes for kit used (e.g. SQK-NBD114-24).
- Trimming of adapters and barcodes will happen by default in Dorado. Once barcodes are trimmed reads can not be demuxed again.
- For more information check the Dorado documentation.
Coverage analysis:
RRMS target bed file can be downloaded from the AS catalogue available here.
Mosdepth is used to check coverage on target regions for the barcodes of interest:
mosdepth -x -t 8 -n -b {target_bed} {out_prefix} {input_pass_bam}
Modification calling
Human variation pipeline is used to aggregate modifications per genomic positions using modkit.
The workflow is available in the following repository: wf-human-variation github.
The documentation can be found in the following space: wf-human-variation EPI2ME page
For most RRMS runs we recommend running the following command:
nextflow run https://github.com/epi2me-labs/wf-human-variation \
-profile singularity \
--mod \
--bam <bam> \
--bed RRMS_human_hg38.bed \
--ref GCA_000001405.15_GRCh38_no_alt_analysis_set.fasta \
--sample_name <sample> --out_dir <output_dir>
(Optional) For haplotype-specific methylation:
If haplotype-specific methylation is required, you can provide options “--snp –phased“ to aggregate modifications identified on each of the haplotypes (i.e. one bedmethyl file for each of the haplotypes will be generated):
nextflow run https://github.com/epi2me-labs/wf-human-variation \
-profile singularity \
--mod --snp --phased \
--bam <bam> \
--bed RRMS_human_hg38.bed \
--ref GCA_000001405.15_GRCh38_no_alt_analysis_set.fasta \
--sample_name <sample> --out_dir <output_dir>
Note: For this specific analysis, a sample coverage of >30X is recommended.
Differentially methylated regions detection:
For detection of differentially methylated regions across different samples “modkit dmr” can be used.
For more information check the modkit documentation available here.
Visualisation:
The BAM file(s) generated by Dorado contains canonical bases as well as per-read modifications stored in MM and ML BAM tags. To visualise the per-read modification calls, IGV can be used to load the BAM file and set "colour reads as" to “base modification 2-color (all)”.
If phasing was performed using wf-human-variation pipeline, the haplotagged BAM file can be uploaded in IGV and alignments can be grouped by haplotype using the IGV option “group by” and selecting “phase”.
Per-position methylation frequencies can also be visualised in IGV by using BIGWIG format. For this, modkit is used to generate BEDGRAPH files using the following command:
modkit pileup --cpg --combine-strands --bedgraph \
--threads 10 --prefix {out_prefix} \
--ref {reference_fasta} \
{out_folder} {input_pass_bam}
Please note, a different bedgraph file will be created for each of the modifications present, in this case 5mC and 5hmC.
Next, bedGraphToBigWig is used to generate bigwig files which can be uploaded together with your BAM file in IGV:
bedtools sort -i {out_folder}/{prefix}_m_CG0_combined.bedgraph | cut -f 1-4 > {out_folder}/{prefix}_m_CG0_combined_sort.bedgraph
bedGraphToBigWig {out_folder}/{prefix}_m_CG0_combined_sort.bedgraph {reference_chrSize} {out_mod_bed_agg_filt_bigwig}
Benchmarking results:
For information about benchmarking the performance of RRMS for human samples, please see our RRMS performance document.
13. Flow cell reuse and returns
We do not recommend washing and reusing your flow cells for this method.
Due to the extended sequencing time, and the multiple flow cell washes and library reloads, we do not recommend re-using the flow cells used in this method.
Re-using these flow cells for subsequent sequencing experiments may result in insufficient data generation for analysis.
Follow the returns procedure to send back flow cells to Oxford Nanopore for recycling.
Instructions for returning flow cells can be found here.
If you encounter issues or have questions about your sequencing experiment, please refer to the Troubleshooting Guide that can be found in the online version of this protocol.
14. Issues during DNA extraction and library preparation
Below is a list of the most commonly encountered issues, with some suggested causes and solutions.
We also have an FAQ section available on the Nanopore Community Support section.
If you have tried our suggested solutions and the issue still persists, please contact Technical Support via email (support@nanoporetech.com) or via LiveChat in the Nanopore Community.
Low sample quality
| Observation | Possible cause | Comments and actions |
|---|---|---|
| Low DNA purity (Nanodrop reading for DNA OD 260/280 is <1.8 and OD 260/230 is <2.0–2.2) | The DNA extraction method does not provide the required purity | The effects of contaminants are shown in the Contaminants document. Please try an alternative extraction method that does not result in contaminant carryover. Consider performing an additional SPRI clean-up step. |
Low DNA recovery after AMPure bead clean-up
| Observation | Possible cause | Comments and actions |
|---|---|---|
| Low recovery | DNA loss due to a lower than intended AMPure beads-to-sample ratio | 1. AMPure beads settle quickly, so ensure they are well resuspended before adding them to the sample. 2. When the AMPure beads-to-sample ratio is lower than 0.4:1, DNA fragments of any size will be lost during the clean-up. |
| Low recovery | DNA fragments are shorter than expected | The lower the AMPure beads-to-sample ratio, the more stringent the selection against short fragments. Please always determine the input DNA length on an agarose gel (or other gel electrophoresis methods) and then calculate the appropriate amount of AMPure beads to use. 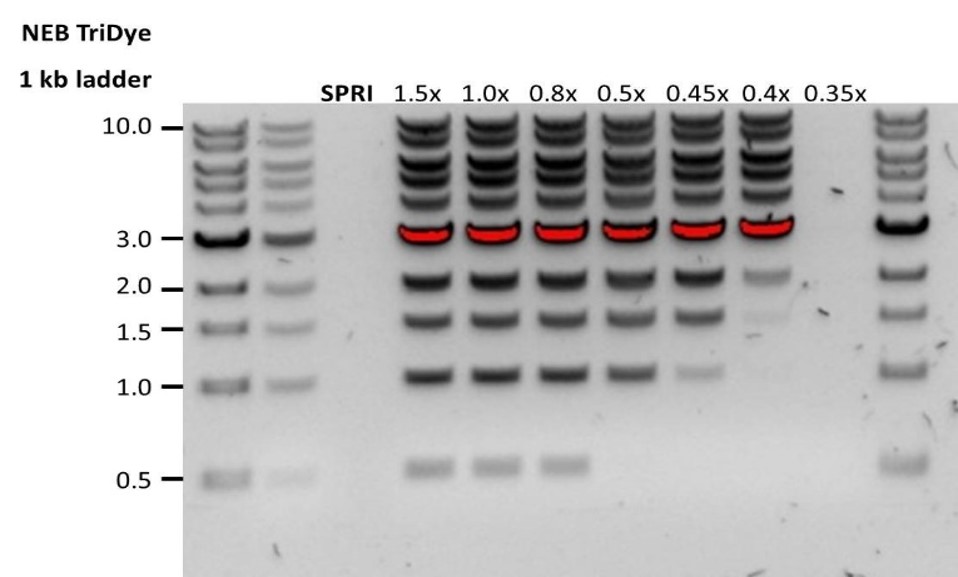 |
| Low recovery after end-prep | The wash step used ethanol <70% | DNA will be eluted from the beads when using ethanol <70%. Make sure to use the correct percentage. |
15. Issues during the sequencing run
Below is a list of the most commonly encountered issues, with some suggested causes and solutions.
We also have an FAQ section available on the Nanopore Community Support section.
If you have tried our suggested solutions and the issue still persists, please contact Technical Support via email (support@nanoporetech.com) or via LiveChat in the Nanopore Community.
Fewer pores at the start of sequencing than after Flow Cell Check
| Observation | Possible cause | Comments and actions |
|---|---|---|
| MinKNOW reported a lower number of pores at the start of sequencing than the number reported by the Flow Cell Check | An air bubble was introduced into the nanopore array | After the Flow Cell Check it is essential to remove any air bubbles near the priming port before priming the flow cell. If not removed, the air bubble can travel to the nanopore array and irreversibly damage the nanopores that have been exposed to air. The best practice to prevent this from happening is demonstrated in this video for how to load a PromethION Flow Cell. |
| MinKNOW reported a lower number of pores at the start of sequencing than the number reported by the Flow Cell Check | The flow cell is not correctly inserted into the device | Stop the sequencing run, remove the flow cell from the sequencing device and insert it again, checking that the flow cell is firmly seated in the device and that it has reached the target temperature. If applicable, try a different position on the device (GridION/PromethION). |
| MinKNOW reported a lower number of pores at the start of sequencing than the number reported by the Flow Cell Check | Contaminations in the library damaged or blocked the pores | The pore count during the Flow Cell Check is performed using the QC DNA molecules present in the flow cell storage buffer. At the start of sequencing, the library itself is used to estimate the number of active pores. Because of this, variability of about 10% in the number of pores is expected. A significantly lower pore count reported at the start of sequencing can be due to contaminants in the library that have damaged the membranes or blocked the pores. Alternative DNA/RNA extraction or purification methods may be needed to improve the purity of the input material. The effects of contaminants are shown in the Contaminants Know-how piece. Please try an alternative extraction method that does not result in contaminant carryover. |
MinKNOW script failed
| Observation | Possible cause | Comments and actions |
|---|---|---|
| MinKNOW shows "Script failed" | Restart the computer and then restart MinKNOW. If the issue persists, please collect the MinKNOW log files and contact Technical Support. If you do not have another sequencing device available, we recommend storing the flow cell and the loaded library at 4°C and contact Technical Support for further storage guidance. |
Pore occupancy below 40%
| Observation | Possible cause | Comments and actions |
|---|---|---|
| Pore occupancy <40% | Not enough library was loaded on the flow cell | Ensure you load the recommended amount of good quality library in the relevant library prep protocol onto your flow cell. Please quantify the library before loading and calculate mols using tools like the Promega Biomath Calculator, choosing "dsDNA: µg to pmol" |
| Pore occupancy close to 0 | The Ligation Sequencing Kit was used, and sequencing adapters did not ligate to the DNA | Make sure to use the NEBNext Quick Ligation Module (E6056) and Oxford Nanopore Technologies Ligation Buffer (LNB, provided in the sequencing kit) at the sequencing adapter ligation step, and use the correct amount of each reagent. A Lambda control library can be prepared to test the integrity of the third-party reagents. |
| Pore occupancy close to 0 | The Ligation Sequencing Kit was used, and ethanol was used instead of LFB or SFB at the wash step after sequencing adapter ligation | Ethanol can denature the motor protein on the sequencing adapters. Make sure the LFB or SFB buffer was used after ligation of sequencing adapters. |
| Pore occupancy close to 0 | No tether on the flow cell | Tethers are adding during flow cell priming (FLT/FCT tube). Make sure FLT/FCT was added to FB/FCF before priming. |
Shorter than expected read length
| Observation | Possible cause | Comments and actions |
|---|---|---|
| Shorter than expected read length | Unwanted fragmentation of DNA sample | Read length reflects input DNA fragment length. Input DNA can be fragmented during extraction and library prep. 1. Please review the Extraction Methods in the Nanopore Community for best practice for extraction. 2. Visualise the input DNA fragment length distribution on an agarose gel before proceeding to the library prep. 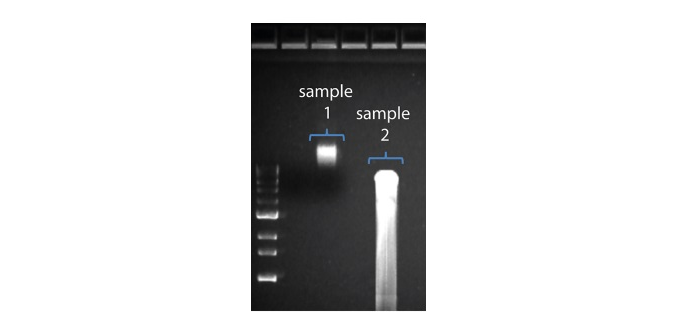 In the image above, Sample 1 is of high molecular weight, whereas Sample 2 has been fragmented. In the image above, Sample 1 is of high molecular weight, whereas Sample 2 has been fragmented.3. During library prep, avoid pipetting and vortexing when mixing reagents. Flicking or inverting the tube is sufficient. |
Large proportion of unavailable pores
| Observation | Possible cause | Comments and actions |
|---|---|---|
Large proportion of unavailable pores (shown as blue in the channels panel and pore activity plot) 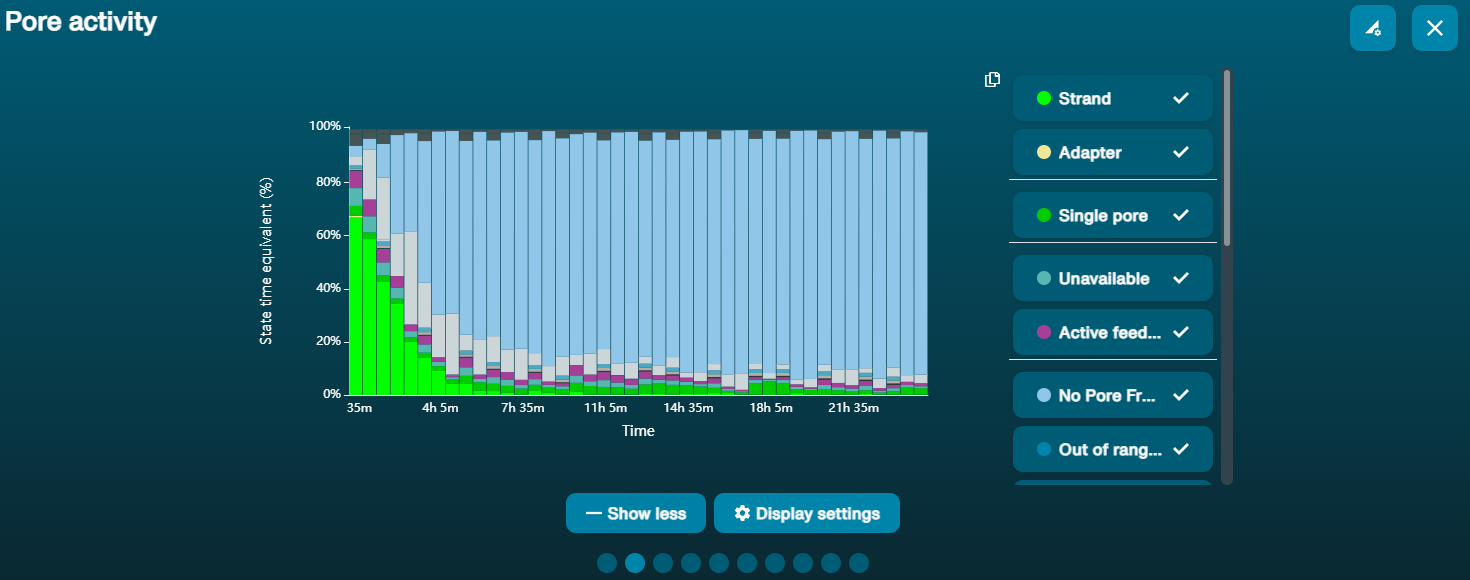 The pore activity plot above shows an increasing proportion of "unavailable" pores over time. The pore activity plot above shows an increasing proportion of "unavailable" pores over time. | Contaminants are present in the sample | Some contaminants can be cleared from the pores by the unblocking function built into MinKNOW. If this is successful, the pore status will change to "sequencing pore". If the portion of unavailable pores stays large or increases: 1. A nuclease flush using the Flow Cell Wash Kit (EXP-WSH004) can be performed, or 2. Run several cycles of PCR to try and dilute any contaminants that may be causing problems. |
Large proportion of inactive pores
| Observation | Possible cause | Comments and actions |
|---|---|---|
| Large proportion of inactive/unavailable pores (shown as light blue in the channels panel and pore activity plot. Pores or membranes are irreversibly damaged) | Air bubbles have been introduced into the flow cell | Air bubbles introduced through flow cell priming and library loading can irreversibly damage the pores. Watch the how to load a PromethION Flow Cell video for best practice. |
| Large proportion of inactive/unavailable pores | Certain compounds co-purified with DNA | Known compounds include polysaccharides. 1. Clean-up using the QIAGEN PowerClean Pro kit. 2. Perform a whole genome amplification with the original gDNA sample using the QIAGEN REPLI-g kit. |
| Large proportion of inactive/unavailable pores | Contaminants are present in the sample | The effects of contaminants are shown in the Contaminants Know-how piece. Please try an alternative extraction method that does not result in contaminant carryover. |
Temperature fluctuation
| Observation | Possible cause | Comments and actions |
|---|---|---|
| Temperature fluctuation | The flow cell has lost contact with the device | Check that there is a heat pad covering the metal plate on the back of the flow cell. Re-insert the flow cell and press it down to make sure the connector pins are firmly in contact with the device. If the problem persists, please contact Technical Services. |
Failed to reach target temperature
| Observation | Possible cause | Comments and actions |
|---|---|---|
| MinKNOW shows "Failed to reach target temperature" | The instrument was placed in a location that is colder than normal room temperature, or a location with poor ventilation (which leads to the flow cells overheating) | MinKNOW has a default timeframe for the flow cell to reach the target temperature. Once the timeframe is exceeded, an error message will appear and the sequencing experiment will continue. However, sequencing at an incorrect temperature may lead to a decrease in throughput and lower q-scores. Please adjust the location of the sequencing device to ensure that it is placed at room temperature with good ventilation, then re-start the process in MinKNOW. |







