Ligation Sequencing sample input and flow cell loading know-how
Requirements
Ligation Sequencing sample input and flow cell loading know-how
FOR RESEARCH USE ONLY
Summary table

Flow cell loading amount for ligation-based libraries
To maximise sequencing output, the pore activity must be high: pores need to be kept filled with DNA, minimising the time that they are idle in between strands. This means maintaining a high pore occupancy, ensuring that as many pores as possible are engaged with DNA at any given time. The benefit is twofold:
- More data is produced per pore per hour.
- The pore retention across the run is higher, resulting in more output from a flow cell.
We have found that achieving high pore occupancy with R10.4.1 Flow Cells (MinION and PromethION Flow Cells) requires ~35–50 fmol of 'good quality library'.
A 'good quality library' can be attributed to the extent of the ligation of sequencing adapters during the library preparation. The presence of sequencing adapters at each end of the DNA molecules is the optimal conformation for tethering to the membrane and subsequent capture by the pore. Molecules without a sequencing adapter cannot be sequenced.
When loading a flow cell, the less library you load, the fewer 'pore-threadable ends' are present on the flow cell. Pores will be 'searching' for molecules for longer, and if the pores are not continuously sequencing, then output could be compromised. It is important to note that we do not observe a linear relationship between input onto the flow cell and sequencing yield, but underloading a flow cell can lead to reduced output.
For libraries of 1 kb and below, we recommend an increased library load of 100 fmol of 'good quality library' (see Figure 1), which requires starting with 200 fmol of input material. More DNA is required on the flow cell in this instance because short fragments have a faster turnover sequencing time than longer ones.
For libraries of 1–10 kb, we recommend starting with 100–200 fmol of input material to ensure you have sufficient library to load (i.e. 35–50 fmol) (see Figure 1).
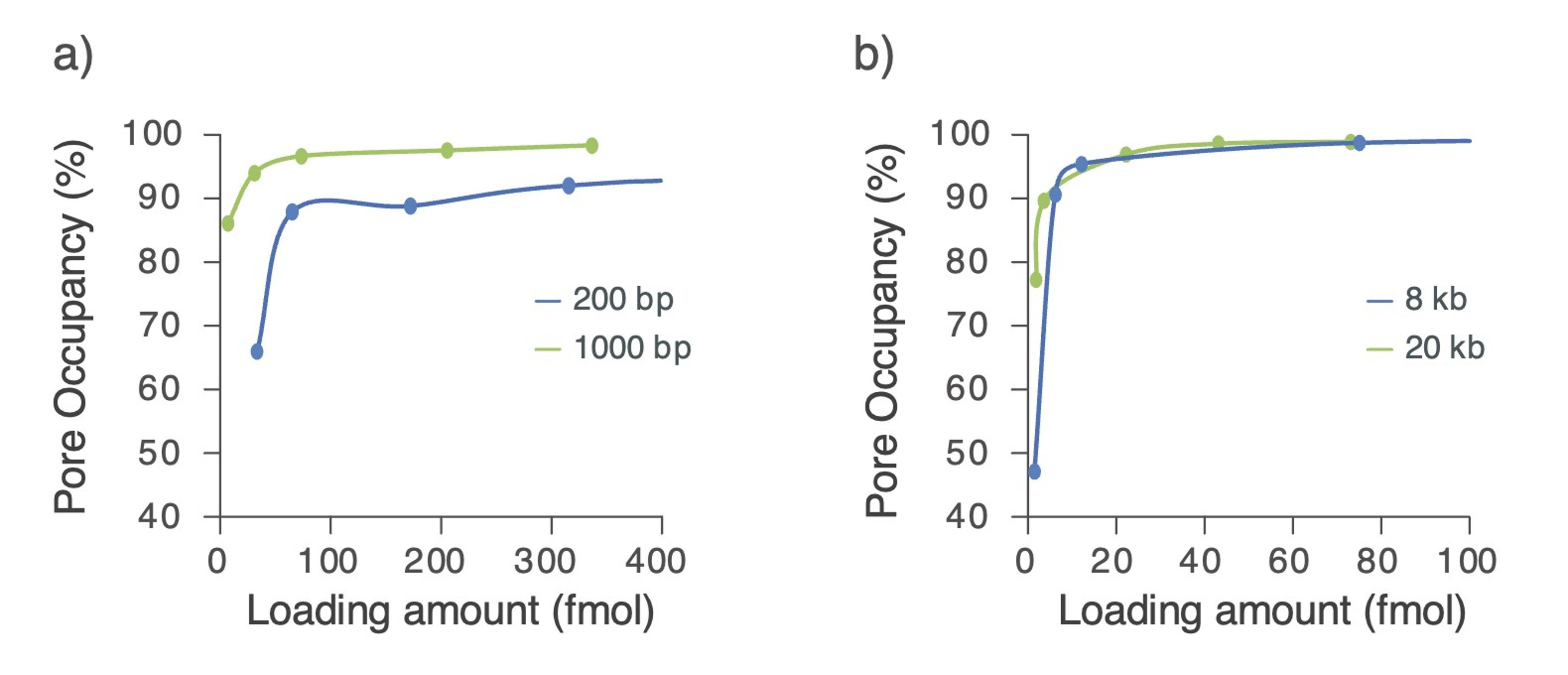
Figure 1. The relationship between the amount of library loaded onto an Oxford Nanopore flow cell and the resulting pore occupancy obtained.
a) For very short libraries (<1 kb), we find that pore occupancy is maximised when 100 fmol of library is loaded. b) We find that pore occupancy is maximised when >35 fmol of 'good quality library' is loaded. Libraries were prepared using the Ligation Sequencing Kit V14 (SQK-LSK114) and loaded onto R10.4.1 Flow Cells.
For high-molecular-weight (HMW), long fragment DNA libraries (>10 kb), we recommend using mass rather than molarity for sample input and flow cell loading. We find that molar quantification of such samples is more difficult and unreliable. For long fragment libraries (>10 kb), we recommend using a starting input of 1 µg. This is almost always sufficient to ensure optimal pore occupancy.
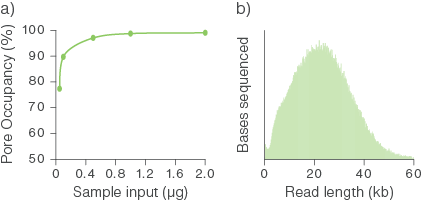
Figure 2. The relationship between HMW DNA input into the library preparation and pore occupancy.
Sequencing libraries were prepared using the Ligation Sequencing Kit with various starting inputs of a gDNA sample comprised of long molecules. All of the yielded libraries were loaded onto PromethION Flow Cells. a) An input of ~0.5–1 µg was sufficient to obtain optimal pore occupancy. b) The fragment length distribution of the library.
Input amount
If you start with less than 1 µg HMW gDNA, then output may decrease as pore occupancy deteriorates. However, even when starting with as little as 100 ng of HMW DNA, we have observed outputs of >50 Gb from PromethION Flow Cells.
As you decrease input to below 100 ng, pore occupancy significantly deteriorates, and we would recommend that users consider shearing the DNA or performing amplification (by PCR or multiple displacement amplification (MDA)) to generate more template.
Shearing HMW templates (e.g. using Covaris g-TUBE or Megaruptor®) increases the number of molecules/ends to thread into the pores, thereby increasing pore occupancy and flow cell output. However, fragmenting the DNA to boost the output means that you are likely to observe a reduction in the observed read length.
We investigated the performance that can be obtained from human gDNA when starting with 100 ng, with and without shearing. It was observed that with shearing (e.g. using Covaris g-TUBE), pore occupancy and output could be increased.
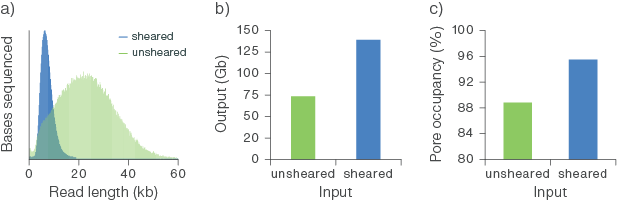
Figure 3. Output on PromethION Flow Cells with 100 ng of HMW human gDNA.
Human gDNA was extracted from whole blood using the QIAGEN Puregene Blood Kit and sheared using Covaris g-TUBE. Libraries were prepared for sequencing with the Ligation Sequencing Kit using 100 ng of sheared and unsheared template DNA. The libraries were run on PromethION Flow Cells. a) Shearing the input gDNA decreases the read lengths observed in the subsequent sequencing. b) The flow cell output (Gb) obtained from 100 ng of input is increased by shearing. c) The shearing of the sample increases the pore occupancy (leading to a higher output from the sheared sample).
Change log
| Date | Version | Change |
|---|---|---|
| 9th January 2026 | v3 | Text was edited to align with writing guidelines and text in graphical images were corrected. |
| 9th May 2025 | v2 | Flongle was removed from document. |
| Aug 2024 | v1 | Initial publication |






