Flongle (FLG_5000_v1_revB_16Oct2018)
TechnicalDocument
Flongle V FLG_5000_v1_revB_16Oct2018
Flongle™ has a disposable flow cell It is low cost It is small-scale It offers potential for pipette-free sample loading Interfaces with our current sequencing platforms
For Research Use Only
Note: This is largely a guide and is subject to change as Flongle evolves rapidly over this development period - please check this page regularly to ensure the instructions provided are correct.
FOR RESEARCH USE ONLY
Contents
What is it?
How it works
What is required
Overview
Flongle™ has a disposable flow cell It is low cost It is small-scale It offers potential for pipette-free sample loading Interfaces with our current sequencing platforms
For Research Use Only
Note: This is largely a guide and is subject to change as Flongle evolves rapidly over this development period - please check this page regularly to ensure the instructions provided are correct.
1. Flongle: The flow cell dongle
Introducing Flongle
Flongle™ (Flow Cell Dongle) is a disposable flow cell with an adapter that provides pipette-free sample application; it is low cost and is a smaller-scale solution to real-time DNA sequencing than a MinION™Mk1B and Mk1C/GridION™/PromethION™ flow cell.

Figure 1. The three main components of the sequencing equipment. The Flongle flow cell lies on top (first tier), supported by the Flongle adapter (second tier), which itself interfaces with the MinION Mk1B (third tier) or GridION (not shown).
It comprises a disposable flow cell containing the biological and chemical elements of nanopore sequencing. The reusable adapter contains a processor (Application Specific Integrated Circuitry (ASIC)) for processing the changes in current in each well, and provides an interface for use with a MinION Mk1B and/or GridION (Figure 1.)
2. The Flongle flow cell
Loading a sample on a Flongle flow cell
The Flongle has the potential of pipette-free sample loading of the prepared library onto the pore-embedded wells via a sample inlet port. The other port available to the user is a waste removal port; a third enables air venting and cannot be accessed. The sample loading port above the Nanopore array allows 30 µl of DNA library to be loaded into the sample reservoir (Fig. 2, green). Excess sample flows into the waste reservoir (Fig. 2, blue), which connects to the waste port. The waste reservoir has a maximum volume of 165 µl and should not be completely filled.
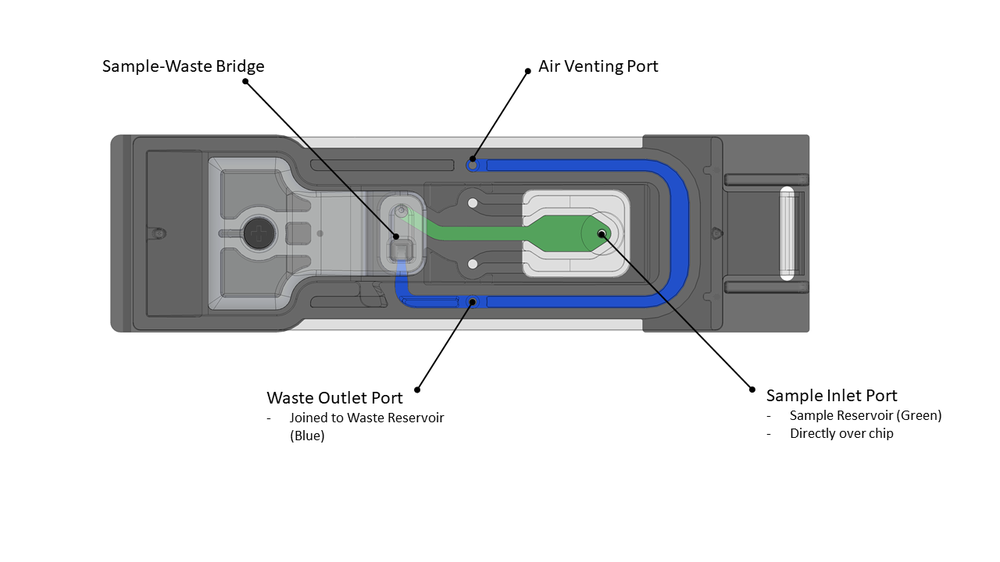
Figure 2. The Flongle flow cell. In green, is the sample reservoir (surrounding the sample inlet port) and, in blue, is the waste reservoir, linking the sample reservoir to the air vent port and waste outlet port (all circled in blue).
For details on how to run a SQK-LSK109 kit on a Flongle please see the SQK-LSK109 protocol for Flongle, here.
Flongle Configuration Test Cell (CTC)
Configuration is the process of testing that communication between the sequencing device and the control software on the host computer is operational prior to experimental work being performed. This is carried out in the absence of any chemistry and uses a specific flow cell known as the Configuration Test Cell (CTC).
3. The Flongle adapter
The Flongle adapter
The Flongle adapter mimics the basal layer of the MinION flow cell and enables disposable, low-cost application of single-use Flongle flow cells. It contains an ASIC, flow cell housing and 126-pin adapter for chip contact and feedback to the ASIC. There is a 32 pin connector to the MinION Mk1B for electrical contact. Figure 3 shows how the Flongle flow cell inserts into the Flongle adapter, attached to a MinION Mk1B.
Figure 3. The Flongle flow cell inserting into the Flongle adapter, attached to a MinION Mk1B.
4. The Flongle sequencing platforms
The MinION Mk1B/GridION interface with a Flongle
The adapter's ASIC enables temperature ramping to 36° C, the optimum for the biological components of the system, via control of the MinION Mk1B's fan or GridION's peltier control. The interface between the flow cell with adapter, and adapter with the MinION Mk1B or GridION, ensure robust connectivity and data collection by the ASIC.
The grounding and shielding of the Flongle through a MinION Mk1B to ensure minimal susceptibility to electromagnetic interference, with maximal sensitivity of the signal-to-noise of current changes.
The circuitry of Flongle (in the adapter) is powered by the MinION Mk1B or GridION. For their Technical Documents, please see below:
5. Hardware for Flongle
Flongle with MinION Mk1B on MinIT
MinIT is an advanced, preconfigured solution to IT infrastructure requirements for running a MinION Mk1B with a Flongle or a regular MinION flow cell.
It can be linked to a laptop, Smartphone or tablet to control the installed MinKNOW software; this enables control of a Flongle and an attached MinION Mk1B.
A link to the MinIT technical document.
Flongle on MinION Mk1B with a laptop
Flongle can be run on a MinION Mk1B, which is subsequently operated by a laptop/computer or MinIT.
The hardware requirements can be found in the MinION and flow cells technical document.
Flongle on a GridION X5
GridION X5 is a sequencing platform that interfaces with up to five Flongle devices, which enables a theoretical collection of 5 Gb in over a 24 hour run time.
6. Software for Flongle
MinKNOW for Flongle
Flongle can be ran on two of our existing sequencing platforms, namely MinION Mk1B and GridION and, as a result, MinKNOW and its various elements are the software that controls, presents and interprets the metrics of a sequencing run.
Guppy is the basecalling element of the MinKNOW package and it directly reads the base sequence from changes in current flow in the flow cell, which are digitised by the ASIC in the Flongle adapter and the circuitry in the MinION Mk1B or GridION. The basecalled sequence is exported by MinKNOW to FASTQ and/or .fast5 files, which enables downstream bioinformatic analysis.
A link to the MinKNOW Technical Document is provided.






