Automated and scalable DNA shearing with the Hamilton STAR for Oxford Nanopore Technologies sequencing
Extraction Method
Automated and scalable DNA shearing with the Hamilton STAR for Oxford Nanopore Technologies sequencing
FOR RESEARCH USE ONLY
Contents
Introduction
Materials
Method
Results
Discussion
Change log
Introduction
For many next-generation sequencing (NGS) applications — including whole genome sequencing (WGS) and structural variant (SV) detection — fragmentation of genomic DNA (gDNA) is a critical step for sequencing success. At scale, this typically requires dedicated hardware, separate from liquid handling systems.
Here, we describe a high-throughput method to mechanically shear gDNA directly on the deck of the Hamilton STAR, using a “tip-shearing” approach. In this method, gDNA in solution is aspirated and dispensed at speed through standard pipette tips, eliminating the need for additional equipment.
With the ability of Oxford Nanopore sequencing to read any fragment length, size selection — often a bottleneck in scalable NGS workflows — is not a required step, but remains optional for tuning fragment size distributions.
The parameters presented here achieve fragment distributions with N50 values of 20+ kb. Further optimisation may be required to target alternative fragment sizes. As with any shearing method, the quality of the input gDNA significantly impacts outcomes. For details on performance and results, see the Sections below.
Materials
Equipment:
- Hamilton STAR with MPH
- DNA QC equipment, e.g. Agilent Bioanalyzer 2100, or Agilent Femto Pulse
Consumables:
- Abgene™ 96 Well 0.8 ml Polypropylene DeepWell™ (ThermoFisher, AB0859)
- 1000 µl CO-RE® II Disposable Tips (Hamilton, 235905)
Method
An automated shearing program is needed to carry out this protocol. A summary of the required shearing parameters is shown in Table 1.
Adjust the concentration of your gDNA samples to 10 ng/μl in 300 μl with nuclease-free water (TE and low TE buffer can also be used).
Transfer samples into a 96-well, 0.8 ml Abgene plate, starting at A1 and filling down to H1 before moving to the next column. Process 8–96 samples in increments of eight.
Seal and briefly centrifuge to collect samples at the bottom of your plate, ensuring no air bubbles are present.
Load the sample plate onto the Hamilton STAR deck.
Begin the automated shearing program.
Once the program is complete, remove the plate from the deck.
Further sample processing may be required depending on downstream workflows e.g. the sample may need to be concentrated. The use of commercially available SPRI beads, such as Agencourt AMPure XP did not adversely affect fragment lengths and is therefore recommended for this purpose.
The settings for the automated shearing are shown in the Table below.
| Parameter | Setting |
|---|---|
| Concentration (ng/µl) | 5 – 15 |
| Volume (µl) | 300 |
| Plate | 0.8 ml Abgene |
| Tips | 1000 µl CO-RE II conductive filtered tip |
| Run length (min) | 10 |
| Pipetting Speeds (µl/sec) | 500 |
| Mixing cycles | 300 |
Table 1: Overview of the required parameters for automated DNA shearing.
Results
To evaluate the effect of sample concentration on shearing outcomes, freshly extracted human gDNA was diluted to 5, 10, and 15 ng/μl in duplicate and processed using the tip-shearing method described above. Unsheared and sheared samples were analysed using the Agilent Femto Pulse System (Figure 1). The traces showed that a lower input concentration led to greater shifts toward lower molecular weights, indicating an inverse correlation between sample concentration and shearing rate.
An additional 20 ng/μl sample was processed under the same conditions, but showed minimal shearing, indicating an upper sample concentration limit for these parameters. The effect of buffer type on shearing was also tested using nuclease-free water, TE, and low TE buffer. There were no observable differences detected for the three reagents (data not shown).
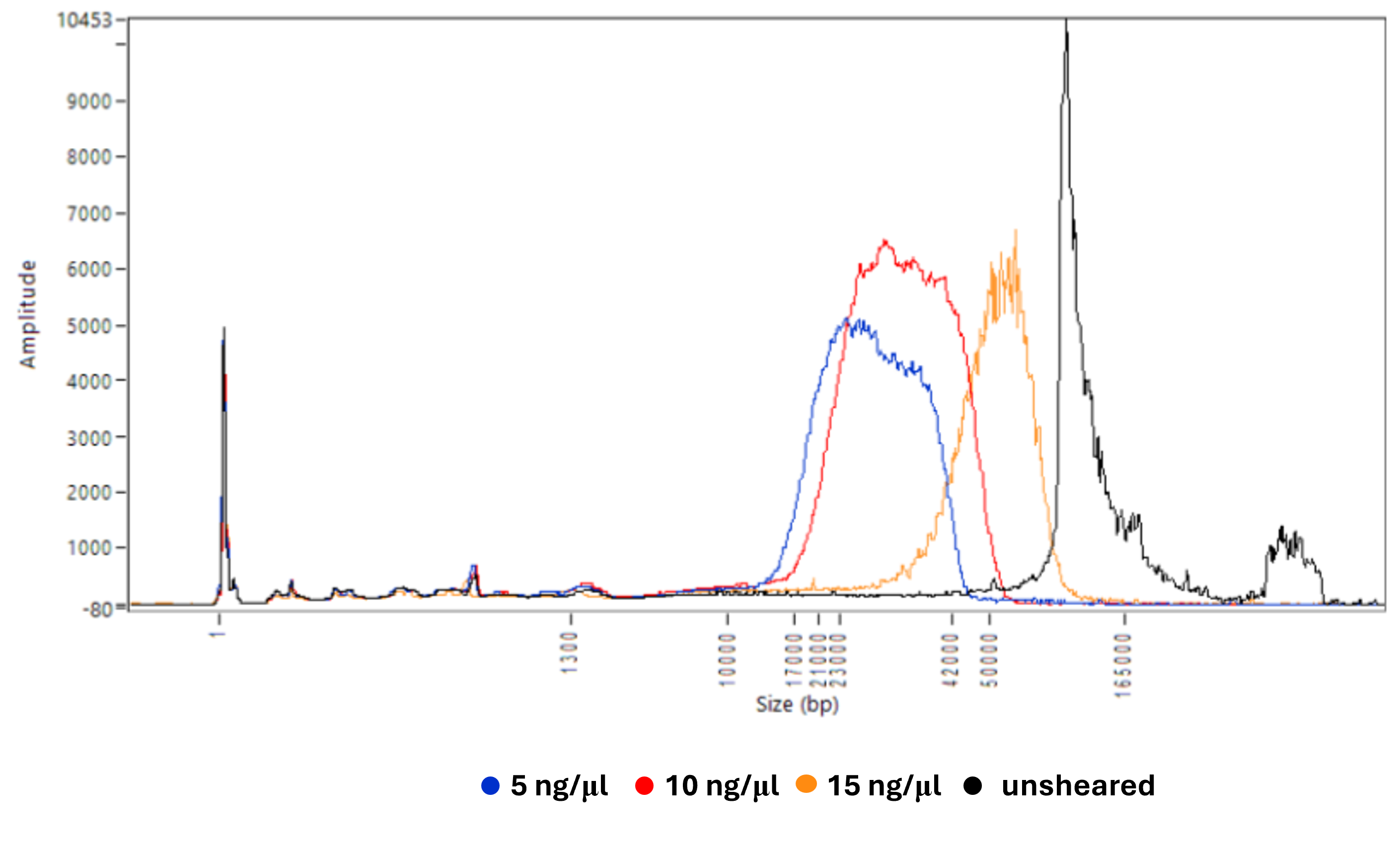 Figure 1: Femto Pulse traces of unsheared and tip-sheared human gDNA at concentrations of 5, 10 or 15 ng/μl on the deck of a Hamilton NGS STAR.
Figure 1: Femto Pulse traces of unsheared and tip-sheared human gDNA at concentrations of 5, 10 or 15 ng/μl on the deck of a Hamilton NGS STAR.
Following shearing, DNA was concentrated using a 1:1 ratio of SPRI beads and eluted in nuclease-free water. Libraries were prepared with the Oxford Nanopore SQK-LSK114 Kit and sequenced for approximately 24 hours using MinION™ Flow Cells on the GridION™ device (see Figures 2 and 3).
For samples diluted to 5 and 10 ng/μl, N50 values of 20 kb and 24 kb were achieved respectively. Both conditions produced fragment distributions tightly centred around their respective N50s, and sequencing output was significantly higher compared with unsheared controls. In contrast, the 15 ng/μl sample yielded a broader fragment size distribution, indicating partial shearing. Further optimisation is necessary to achieve complete fragmentation at this concentration.
All sheared samples contained a subpopulation of shorter DNA fragments (<10 kb). If required, this population can be reduced by introducing a size selection step to enrich for longer fragments.
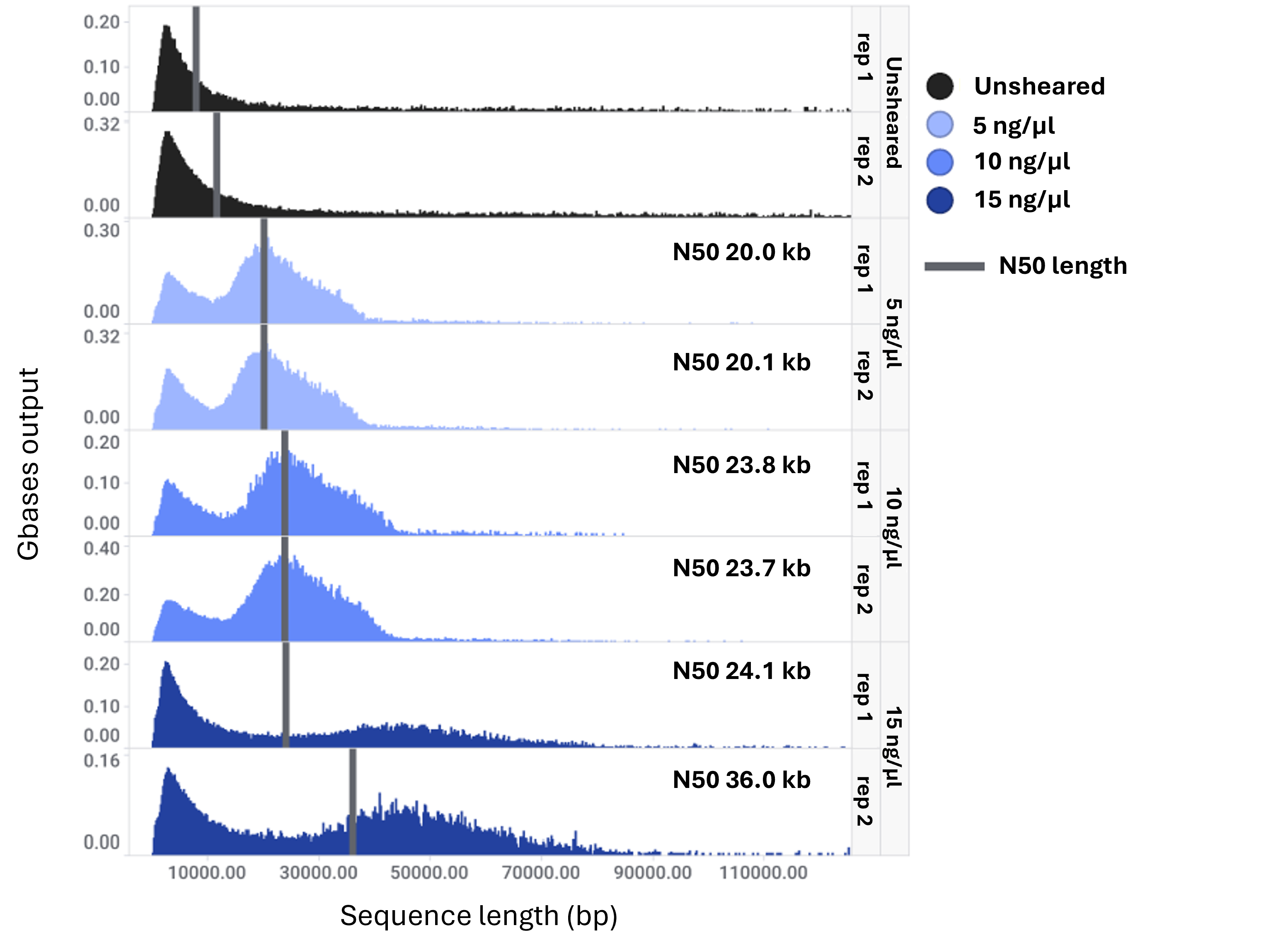
Figure 2: Fragment length distribution histograms of sequenced DNA either sheared at 5, 10 or 15 ng/μl or remained unsheared. The N50 values for sheared samples are shown for each panel.
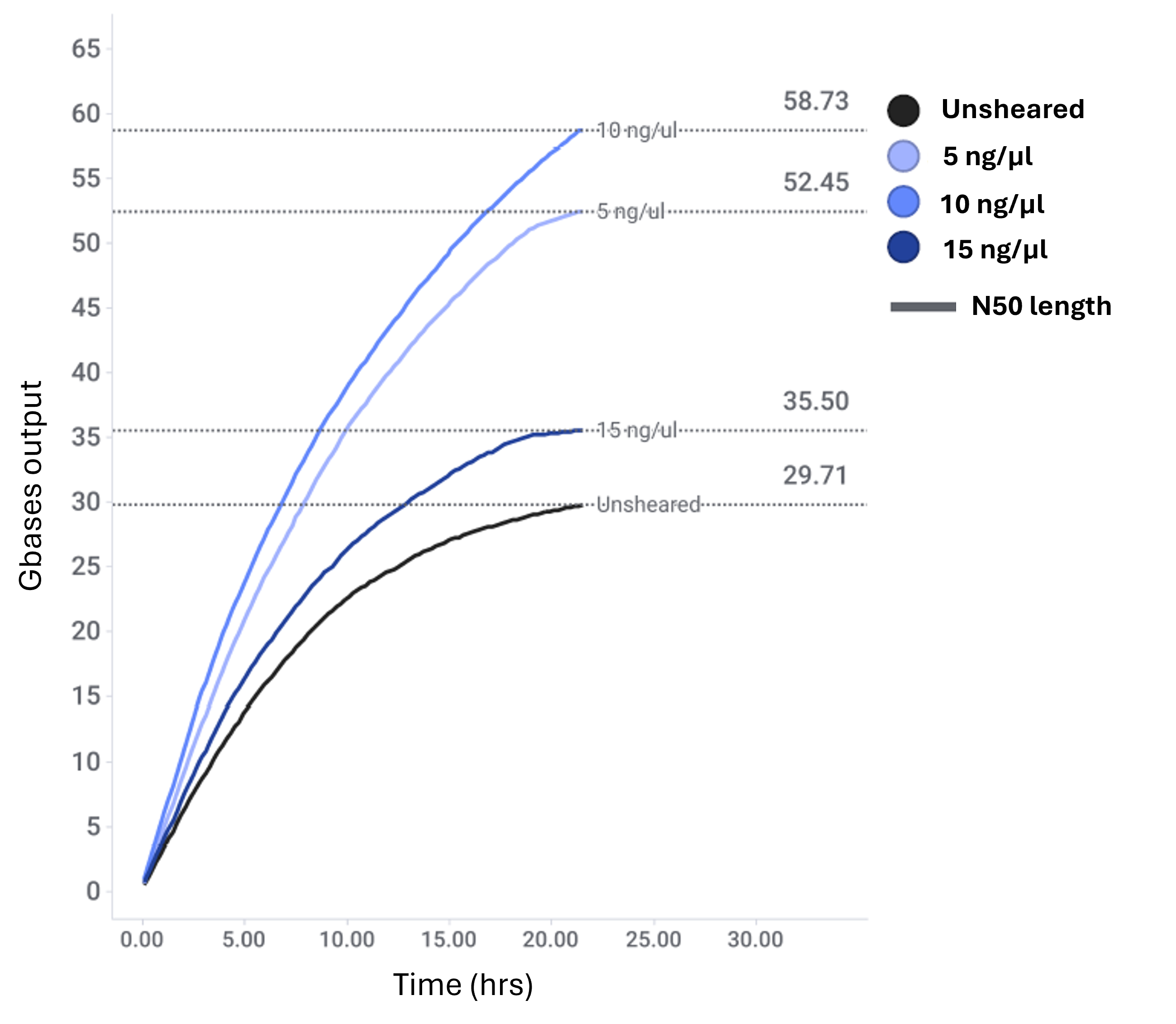
Figure 3: Sequence output line graphs of DNA sequenced for ~ 24 hours showing the increased data output for tip-sheared DNA compared with unsheared DNA.
Discussion
This work demonstrates a scalable and cost-effective method for mechanical shearing of gDNA using liquid handling robots for nanopore sequencing. By leveraging precise pipetting parameters intrinsic to systems like the Hamilton STAR, this tip-shearing approach reduces workflow complexity, time, and capital expenditure.
Fragment length distributions can be further tuned by adjusting shearing parameters. While the protocol was implemented on a Hamilton platform, it is expected to be transferable to other liquid handlers with comparable pipetting capabilities.
This approach complements existing scalable shearing methods (e.g. using GenoGrinder or FastPrep), and offers an alternative that integrates directly into automated NGS workflows.
Change log
| Date | Version | Changes made |
|---|---|---|
| 25th September 2025 | v1 | Document release |






