Q-Line sequencing software user guide (Q_SSU_revB_03Feb2025)
Protocol
Q-Line sequencing software user guide V Q_SSU_revB_03Feb2025
FOR RESEARCH USE ONLY
Contents
Introduction to the Q-Line sequencing software
Log in and launch the application
Create a sample sheet
Set up and run an assay
Run complete
Technical run report
1. Introduction
Introduction to the sequencing software
The sequencing software carries out several core tasks: data acquisition, real-time analysis and feedback, basecalling, data streaming, controlling the device, and ensuring that the platform chemistry is performing correctly to run the samples. The sequencing software takes the raw data and converts it into reads by recognition of the distinctive change in current that occurs when a DNA strand enters and leaves the pore. It then basecalls the reads and writes out the data into FASTQ and BAM files.
Computer specification
Please refer to the Q system GridION IT requirements document for details of the IT setup required to run a GridION Q.
2. Log in and launch the application
Click on the user account. You will be asked for your password.
If you have created other user accounts besides GridION user, you will see them in a list.

You will only see local accounts, not Active Directory (AD) accounts. To log in using an AD account, click “Not listed?".

Then, enter the account name without the domain name (e.g., if the domain account name is John.Smith@example.local, enter John.Smith in the username prompt).
Note: If you are rebooting your device, you will be asked for your device passphrase, which differs from your user account password. For more information on how to set up your user account and change your device passphrase, see the "User management - local user" of the Q-line software installation and maintenance guide.
Enter the password. If you have not changed the password for GridION user, the password will be “grid“.

In the sidebar, click the Sequencing Software icon.

Once you open the Sequencing Software, the Overview page will be displayed. The overview shows the five flow cell positions and their current status.

From this page, you can start running an assay.
There are four buttons on the left side:
- Overview: The overview includes the five GridION positions and their current status. From this page, it is possible to set up an assay through the Set up run buttons.
- Run setup: The run setup opens to the assay selection. It shows the five stages of setting up a run.

- Run log: The run log contains all your completed runs.

- System: This section includes information on device storage and language settings. If you have Laboratory Manager or IT Administrator permissions, you will also see Assay Configuration and API Key Management.

From this page, you can begin the following tasks: set up a run, view in-progress or completed runs, manage the device, or view details for live positions.
There are four buttons on the left side:
Overview
The overview includes the five GridION positions, and you can interact on this page to set up your experiment.
Run Setup
Run setup opens up to the assay selection. You can see the five run setups available: Assay selection, flow cell check, sample sheet, prime and load, review and start.
Run log
The run log contains all of the runs you have completed.

System
This section includes: Access device information, Language settings and, if you have Laboratory Manager or IT Administrator permissions, Assay configuration and API key management.

3. Changing your password
Search “Settings” in the search bar and click on Settings.

View the GridION username and the current password by clicking the chevron next to the password.

Change your password by entering your current and new passwords in the relevant boxes. Make sure you follow the password requirements listed below.

Password requirements
Note: IT Administrators will be able to change these complexity requirements.
- You can use numbers, letters, upper/lower case and punctuation.
- The minimum password length is eight characters.
- You cannot have a password that is a palindrome (reads the same backwards and forwards, e.g. “racecar").
- The new password cannot be the same as the previous one, even if you have changed the cases.
- The new password cannot be the rotated version of the old password (e.g. "billy" and "illyb").
- Multiple character groups must be included in the new password.
- The password must contains at least one character from three of four character groups. These groups are:
- Digits
- Lower case alphabetic characters from the current system locale
- Upper case alphabetic characters from the current system locale
- Other characters, e.g. punctuation marks and symbols such as * % ! ?
For local users where there is no distinction between upper and lower case (i.e. most Asian scripts), the two groups are treated as one.
You can also click the button next to New Password to create a strong password.


4. Create a sample sheet
When setting up an assay run, you will need to provide a sample sheet detailing the barcodes used, the corresponding sample IDs, and their sample type. This is a vital traceability link, and strict validation rules are applied to minimise the risk of data entry errors.
You can manually create sample sheets on the device using LibreOffice Calc or text editor, available on the GridION. You can also create the sample sheet off-device and transfer it onto the device through drive mounting (to do this, speak to your IT department). You can also transfer your sample sheets via a USB stick (to do this, enable the USB mount as described in the "Managing data" section of the Q-Line Sequencing Software installation and maintenance guide.
Alternatively, you can use a laboratory information management system (LIMS) to add information to your sample sheet automatically. The GridION and LIMS must be configured before library preparation, as detailed in the “Configuring LIMS integrations” section of the Q-Line Sequencing Software installation and maintenance guide.
Manual input
For laboratories without a LIMS, sample sheets can be written manually using Microsoft Excel, LibreOffice Calc, or a text editor. Whichever is used, it is vital to save the file as in the .csv format.
LibreOffice Calc is installed on your GridION. The following steps describe how to use it to generate a sample sheet.
Open the template for the assay sheet in LibreOffice Calc.
Edit the template for the samples you are running.
Save the sample sheet as a .csv file (not ODS template) in your local folder.
Import the sample sheet when setting up an assay, as described in the "Set up and run and assay" section of this guide.
Template format: available as templates (.ods files) in your GridION
| Keys | Description |
|---|---|
v1 (above header section) | Version of the sample sheet format (Do not change this) |
assay | Assay key installed on the UI and its version. Must be in the format key:version where key and version are defined in the assay definition file |
library_id | Provided by you to ID the library for sequencing (Any of a-z A-Z 0-9 \.\-_]+[,\s]*$)Important: Do not add any personally-identifiable information in this field. |
sample_count | Number of samples used. This must match the number of barcodes used, including samples and controls. Note: Using the template will automatically calculate this from the barcode/sample section. |
created_by | (Optional) User who prepared the library (Any of a-z A-Z 0-9 \.\-_]+[,\s]*$) |
lot | (Optional) The kind of item, and the unique ID for lot used. (In separate cells on the same row, see image below for an example) |
Barcode/sample section
barcode | The barcode used during library preparation for the sample. Important: If you are not using barcoding, you will not have the barcode column and will only have a sample count of 1. |
sample_id | Provided by you to ID individual input samples. It must be unique within the assay. Important: Do not add any personally-identifiable information in this field. |
sample_type | The type of sample. For more information on acceptable fields, see the "Configuring an assay definition file for your own assay" section of the Q-Line Sequencing Software Installation and maintenance guide. This field will depend on the assay run and its definitions, such as sample and control sample. |
Examples
The table below shows the sample sheet for an assay using the Ligation Sequencing Kit without barcoding. Important note: empty rows below v1 and sample_count rows must be conserved in your own sample sheets. A failure to do so will invalidate the document.
v1 | |
assay | ligation-kit09 |
library_id | My_Library_ID |
sample_count | 1 |
sample_type | sample_id |
sample | Test_01 |
Below is an example sample sheet for the Rapid Barcoding Kit:
v1 | ||
assay | rapid-barcode-96-kit10:v1.0 | |
library_id | My_Library_ID | |
sample_count | 5 | |
barcode | sample_type | sample_id |
barcode01 | no_template_control | Control_01 |
barcode02 | no_template_control | Control_02 |
barcode03 | sample | Test_01 |
barcode04 | sample | Test_02 |
barcode05 | sample | Test_03 |
5. Set up and run an assay
This section explains how to set up and conduct your assay. We will use the Rapid Barcoding Experiment 01-96 assay as an example, and will guide you through selecting your assay, performing a flow cell check, adding a sample sheet, priming and loading your flow cell, and initiating your run.
Click "Overview" to see the five flow cell positions. Then click "Set up run".
You can also click Run setup in the sidebar. Note: The Set up run option is only available if flow cells are inserted and available for use.

Choose the Rapid Barcoding Experiment 01-96 assay, and click Continue.
You can also add custom assays to this list; the method to generate a custom assay is described in the "Configuring an assay definition file for your own assay" of the Q-Line Sequencing Software Installation and maintenance guide.

Click Check flow cell. This will initiate the check for whether the flow cell is usable for the assay.

A status message will appear under the flow cell position: “Checking flow cell...". You can also cancel it by clicking Cancel.

Once the flow cell check has finished, the software will report whether the flow cell has sufficient health to run the assay. Click Continue to move to the next step.
A failed flow cell check will prevent the start of a run. However, it is possible to override a low flow cell health check with certain assays, including most default assays and custom assays if configured that way. If your assay allows the overriding of low flow cell health, the number of available pores is displayed to enable you to make an informed decision.
In some cases, the low flow cell health might have been caused by poor thermal contact between the position and the flow cell. In these cases, on-screen instructions will direct you to reinsert the flow cell and try again.

Select the sample sheet that you want to use. You can only select sample sheets that have a Validation status of Pass. If the sample sheet you want to use has a different status, click View sheet for more information. You may need to use a different flow cell.
Tip: If you have a barcode scanner plugged into the GridION, you can scan a barcode for the Library ID of the sample sheet you want to use and filter the list to only display the matching sample sheet.
If your sample sheet is not already displayed, you can create and upload one. The method for creating a sample sheet can be found in the "Create a sample sheet" section of this guide.
To manually upload your sample sheet, click Import.
The sequencing software will check whether the sample sheet meets the requirements. If no issues have been found, click Add sample sheet.

If your sample sheet has an issue, an error message will pop up, advising you which part of the sample sheet does not match the required criteria.
Occasionally, when adding the sample sheet, the progress bar may not move (as shown in the image below). However, clicking Add sample sheet will enable the sample sheet to be added.

When the sample sheet has been imported, a green tick will show it has passed validation. You can also check your sample sheet by clicking the View sheet button.

When you are happy with your sample sheet selection, click Continue.
Prime and load your flow cell. You can see detailed instructions for this in the View full instructions tab.
Once you have loaded your sample, click Continue.
You can now review the assay, libraryID, number of samples, and time to report and check your sample sheet.
If you are happy with the settings, click Run assay. A small prompt will appear in the bottom right corner stating that the run has started. If you did not mean to start it, click Undo within 30 seconds, and the run will stop.

Click the View run button to see the status of your run.

The View run button will open the Run log screen for a particular position. There, four panel are displayed: Run status, Run summary, Run reports and Samples.
- Run status: displays the Initial health check and the Initial sequencing performance. Both reports on potential issues with the run:
- Initial health check: reports on issues introduced between the flow cell check and the sequencing.
- Initial sequencing performance: reports on potential issue with the library.
- Run summary: recapitulates the configuration of the run.
- Run reports: provides a list of reports available.
- Samples: If you have barcodes, you can also check whether individual samples barcodes are found.
Note that this information is also available in the Overview page.

6. Run complete
After the run finishes, view the run report by clicking Run log. It provides details on the run summary, run status, run reports, and sample information.

Click View under the run reports section to access the technical run report.
The report contains information relating to the sequencing performance of your run, including a variety of graphs and statistics. These are useful for troubleshooting and quality control.

You can save the final technical run report as a HTML file by clicking the Save a copy button in the top right corner of the screen.
Note: Additional reports (in PDF or HTML format) will figure in the Sample ID list if an analysis was available for your assay.
Example: Instrument qualification run report


7. Data location
If data offload is configured, sequencing and analysis data and reports are moved to:
/data/offload/<assay_run_id>/
From here, they are moved to the network drive location set by the IT administrator. For data offload instructions, refer to the “Managing data” section of the Q-Line sequencing software installation and maintenance guide.
Sequencing data and reports are located in:
/data/offload/<assay_run_id>/sequencing
The file structure within the sequencing folder is as follows:
|-- /data/offload/<assay_run_id>/sequencing
| |-- barcode_alignment.tsv
| |-- final_summary.txt
| |-- pore_activity.csv
| |-- report.html
| |-- report.json
| |-- report.md
| |-- sample_sheet.csv
| |-- sequencing_summary.txt
| |-- throughput.csv
| |-- fastq_fail
| | |-- barcode01
| | | |-- <flow_cell_id>_fail_barcode01_<id>.fastq.gz
| | |-- barcode02
| | | |-- <flow_cell_id>_fail_barcode02_<id>.fastq.gz
| | |-- unclassified
| | | |-- <flow_cell_id>_fail_unclassified_<id>.fastq.gz
| |-- fastq_pass
| | |-- barcode01
| | | |-- <flow_cell_id>_pass_barcode01_<id>.fastq.gz
| | |-- barcode02
| | | |-- <flow_cell_id>_pass_barcode02_<id>.fastq.gz
| | |-- unclassified
| | | |-- <flow_cell_id>_pass_unclassified_<id>.fastq.gz
| |-- other_reports
| | |-- pore_scan_data.csv
The output location for analysis data and reports is:
/data/offload/<assay_run_id>/analysis
The file structure within the analysis folder is as follows. There is a set of output files for every sample/replicate run in the assay.
|-- /data/offload/<assay_run_id>/analysis
| |-- <output_file>.vcf
| |-- <output_file>.bam
| |-- <output_file>.bam.bai
| |-- <output_file>.html
| |-- <output_file>.pdf
| |-- execution
| | |-- report.html
| | |-- timeline.html
| | |-- trace.txt
8. Technical run report
Technical run report overview
Run reports contain information about the sequencing run and include performance graphs. These graphs are automatically generated at the end of a run or when you click the View button in the Run report panel of the Run log tab.

The run report includes panels for the Run summary, Run configuration, Sequence output, Alignment (only if live alignment is part of your assay), Run health and Run log. It is interactive: sections of interest can be expanded and navigated while troubleshooting suggestions are made available for performance enhancement.
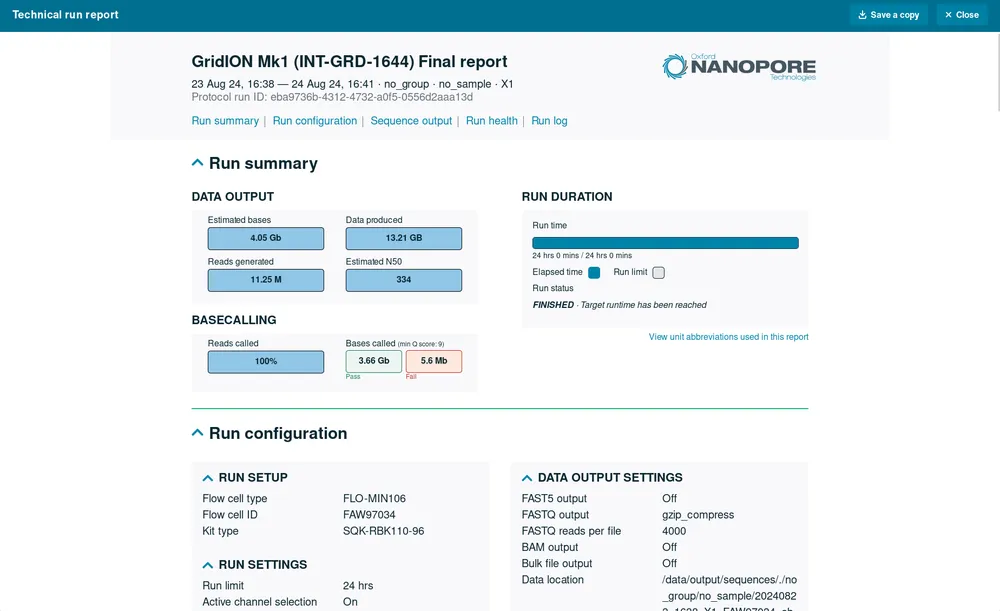
Run summary
This is an overview of the sequencing run, including output and basecalling results and the duration of the run.

Run configuration
A detailed overview of the settings selected for the sequencing run and data output, including the software versions used to sequence the data. It also includes further information about the modified bases and basecalling models used, expansion kits used and read-splitting preferences.

Sequence output
This section includes a more detailed view of the sequencing output, such as read lengths, barcodes detected and quality score. This section of the Technical run report displays Read lengths, Barcodes, Cumulative output and Quality scores data.
Read lengths This graph shows the total number of bases against the read length. Any outliers are displayed in the table below, displaying read length and aggregated reads.

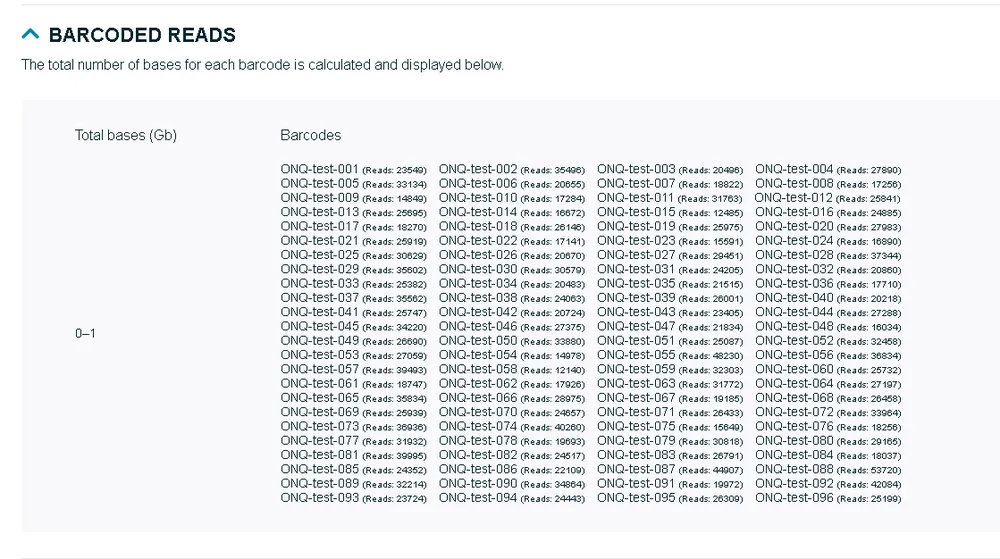
Barcoded reads
This section will be visible only if barcoding is enabled in the assay. It will then show the total number of bases for each barcode.
Cumulative output
The graphs show the total output of the bases and reads sequenced during the experiment.

Quality score
The quality score is calculated as basecalling is performed on your device. Reads that fall below the minimum value of 9 will be classified as failed reads. You can alter the accepted minimum quality score in the assay configuration file.

Alignment
This section will figure in your Technical run report if an alignment file was provided and Alignment enabled in the assay configuration file.
Reference alignment: This section will display the total number of passed reads aligned to each uploaded reference target, calculated and displayed.
Bed regions: If there is corresponding .bed region information for aligned references, it will be shown here.

Run health
This section provides a detailed view of the Pore activity and Pore scan results throughout the run, along with graphs displaying the translocation speed and temperature.



Run log
The run log includes system messages sent during the sequencing run regarding errors, warnings, and any events.







