Flow Cell Wash Kit (EXP-WSH003)
- Home
- Documentation
- Flow Cell Wash Kit (EXP-WSH003)
Protocol
Flow Cell Wash Kit (EXP-WSH003) VWFC_9088_v1_revG_18Sep2019
For Research Use Only
FOR RESEARCH USE ONLY
Contents
Introduction to the protocol
Flushing, reloading or storing a flow cell
Overview
For Research Use Only
1. Overview of the protocol
Introduction to the Wash Kit
The Wash Kit allows sequential runs of multiple sequencing libraries on the same flow cell. It works by washing out the first library, and refreshing the system ready for a subsequent library to be loaded. This procedure provides the opportunity to utilise the same flow cell a number of times, maximising the available run time, particularly for cases where less data per library is required. Following the wash step, Storage Buffer can be introduced into the flow cell, allowing storage of the flow cell before subsequent library additions. The Flow Cell Wash Kit is compatible with R9.4.1 and R10.3 flow cells.
Please note, although the wash procedure should remove 99.9% of the library, some residual DNA may remain on the flow cell. For this reason, users may prefer to barcode their libraries when used in conjunction with the Wash Kit, such that reads from different libraries can be separated from each other. RNA is also efficiently removed, and an RNA barcoding option will become available in the near future. Successful deconvolution of DNA reads has been demonstrated in Oxford Nanopore's internal development:
Figure 1. A) Sequential loading of barcoded libraries without washing. B) Sequential loading of barcoded libraries with washing. With washing, only residual library remained on the flow cell.
For users who wish to use barcoding to run multiple libraries at one time rather than washing the flow cells, please see the barcoding kits we have available:
Nuclease activity of the Flow Cell Wash Kit
The Flow Cell Wash Kit contains DNase I, that is used to digest any remaining DNA library on a flow cell. Once the library is removed, the flow cell can be re-used immediately or stored for later use.
During sequencing, an accumulation of pores in the “unavailable" state (Figure 2) may be observed, causing the rate of data acquisition to decline as fewer pores are available to accept and sequence strands. We have demonstrated that in these circumstances, pores can be reverted to the “active pore” state by pausing sequencing and washing the flow cell with the DNase I in the Flow Cell Wash Kit. In Figure 2, the astrisks indicate where sequencing has been paused and the flow cell washed. Note: If the sequencing run is paused in MinKNOW for the flow cell wash, you will only see the restoration of sequencing pores after a new pore scan has been performed.
The wash step is recommended where sequencing channels are lost to the “unavailable” state (Figure 2). In circumstances where channels have been lost by other means, for example “saturated”, the wash step is not effective at reverting channels to the “active pore” state. If your sample is not known to block and cause saturation of the flow cell, we recommend loading a fresh library on a new flow cell. If you do not have excess library to load on a new flow cell, you can recover the library and reload on a new flow cell following method 1 of the Library recovery from flow cells protocol.
 Figure 2. Pore states observed on a flow cell before and after wash steps are performed. A flow cell has been loaded with a sequencing library that has resulted in an accumulation of pores in the “unavailable” state, leading to a decrease in the rate of data acquisition. The red asterisks indicate when a wash step has been performed. A significant number of the pores that had been lost to the “unavailable” state have reverted to the “Pore available” state and are available for sequencing once again.
Figure 2. Pore states observed on a flow cell before and after wash steps are performed. A flow cell has been loaded with a sequencing library that has resulted in an accumulation of pores in the “unavailable” state, leading to a decrease in the rate of data acquisition. The red asterisks indicate when a wash step has been performed. A significant number of the pores that had been lost to the “unavailable” state have reverted to the “Pore available” state and are available for sequencing once again.
In experiments where output is limited by the increase in pores in the “unavailable” state, we have shown that output can be improved by performing several wash steps over the lifetime of a flow cell. Figure 3 shows the output obtained from a PromethION Flow Cell loaded with a library of DNA extracted from chicken - Gallus gallus, and a MinION Flow Cell loaded with a library of DNA extracted from a type of Japense ricefish - Oryzias latipes, where unavailable pores increased over the course of the experiment, and so flow cell washes were performed to unblock the pores (Figure 3). In each case, the use of multiple washes allowed for an improvement of the output from the flow cell, without any compromise in observed read length (Figure 4).
Figure 3. Throughput observed from Gallus gallus and Oryzias latipes libraries run on a PromethION Flow Cell and a MinION Flow Cell, respectively. The arrows indicate the timing of each wash step: wash steps were performed at the point where the rate of data acquisition started to slow due to the accumulation of “recovering” pores. In each case, output is more than doubled from the point of the first wash.
Figure 4. Effective inactivation of the DNase I prevents read length deterioration in the experiments after a nuclease wash is performed. In this example, the read length of the Gallus gallus library was recorded before the first wash (load #1) and then again after the first and second washes (load #2 and #3, respectively). No decrease in read length is observed.
2. Equipment and consumables
Materials
- Flow Cell Wash Kit (EXP-WSH003)
Equipment
- P1000 pipette and tips
- P20 pipette and tips
- Ice bucket with ice
Flow Cell Wash Kit contents (EXP-WSH003)
| Contents | Volume (µl) | No. of tubes | No. of uses |
|---|---|---|---|
| Wash Solution A | 140 | 1 | 6 |
| Wash Solution B | 1300 | 2 | 6 |
| Storage Buffer | 1600 | 2 | 6 |
- Wash Solution A contains DNase I.
- Wash Solution B contains the exonuclease buffer that maximises activity of the DNase I.
- The Storage Buffer allows flow cells to be stored for extended periods of time.
3. Flushing a MinION/GridION Flow Cell
Materials
- Flow Cell Wash Kit (EXP-WSH003)
Equipment
- P1000 pipette and tips
- P20 pipette and tips
- Ice bucket with ice
Preparation to run the washing procedure
- This protocol assumes that the flow cell has already had a DNA/RNA library run on it
- The aim is to remove most of this initial library from the flow cell
- The Wash Kit contains all solutions required for removal of the initial library
- Data acquisition in MinKNOW should be stopped (if loading a new library or storing the flow cell), or paused (if loading more of the same library after the wash)
- After the flow cell has been washed, a new library can be loaded or the flow cell can be stored at 4°C
Place the tube of Wash Solution A on ice. Do not vortex the tube.
Thaw one tube of Wash Solution B at room temperature.
Mix the contents of Wash Solution B thoroughly by vortexing, spin down briefly and place on ice.
In a clean 1.5 ml Eppendorf DNA LoBind tube, prepare the following Wash Mix:
| Component | Volume |
|---|---|
| Wash Solution A (A) | 20 μl |
| Wash Solution B (B) | 380 μl |
Mix well by pipetting, and place on ice. Do not vortex the tube.
Stop or pause the sequencing experiment in MinKNOW, and leave the flow cell in the device.
Before removing the waste fluid, ensure that the flow cell priming port cover and SpotON sample port cover are closed, as indicated in the figure below.
Remove all fluid from the waste channel through waste port 1 using a P1000 pipette.
As both the flow cell priming port and SpotON sample port are closed, no fluid should leave the sensor array area.
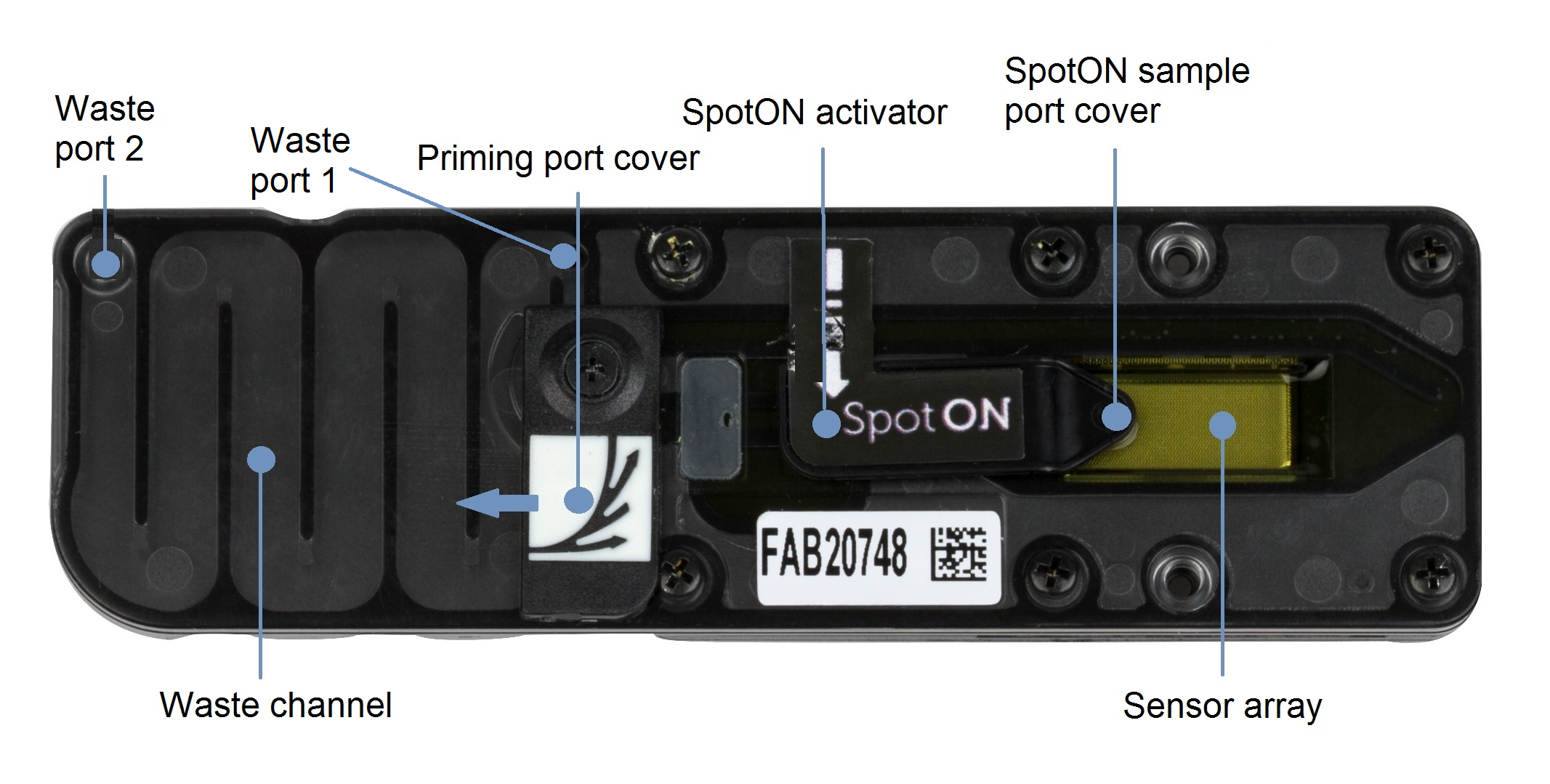
IMPORTANT
It is vital that the flow cell priming port and SpotON sample port are closed before removing the waste buffer to prevent air from being drawn across the sensor array area, which would lead to a significant loss of sequencing channels.
Slide the flow cell priming port cover clockwise to open.
After opening the priming port, check for a small air bubble under the cover. Draw back a small volume to remove any bubbles:
- Set a P1000 pipette to 200 µl.
- Insert the tip into the flow cell priming port.
- Turn the wheel until the dial shows 220-230 µl, or until you can see a small volume of buffer/liquid entering the pipette tip.
- Visually check that there is continuous buffer from the flow cell priming port across the sensor array.
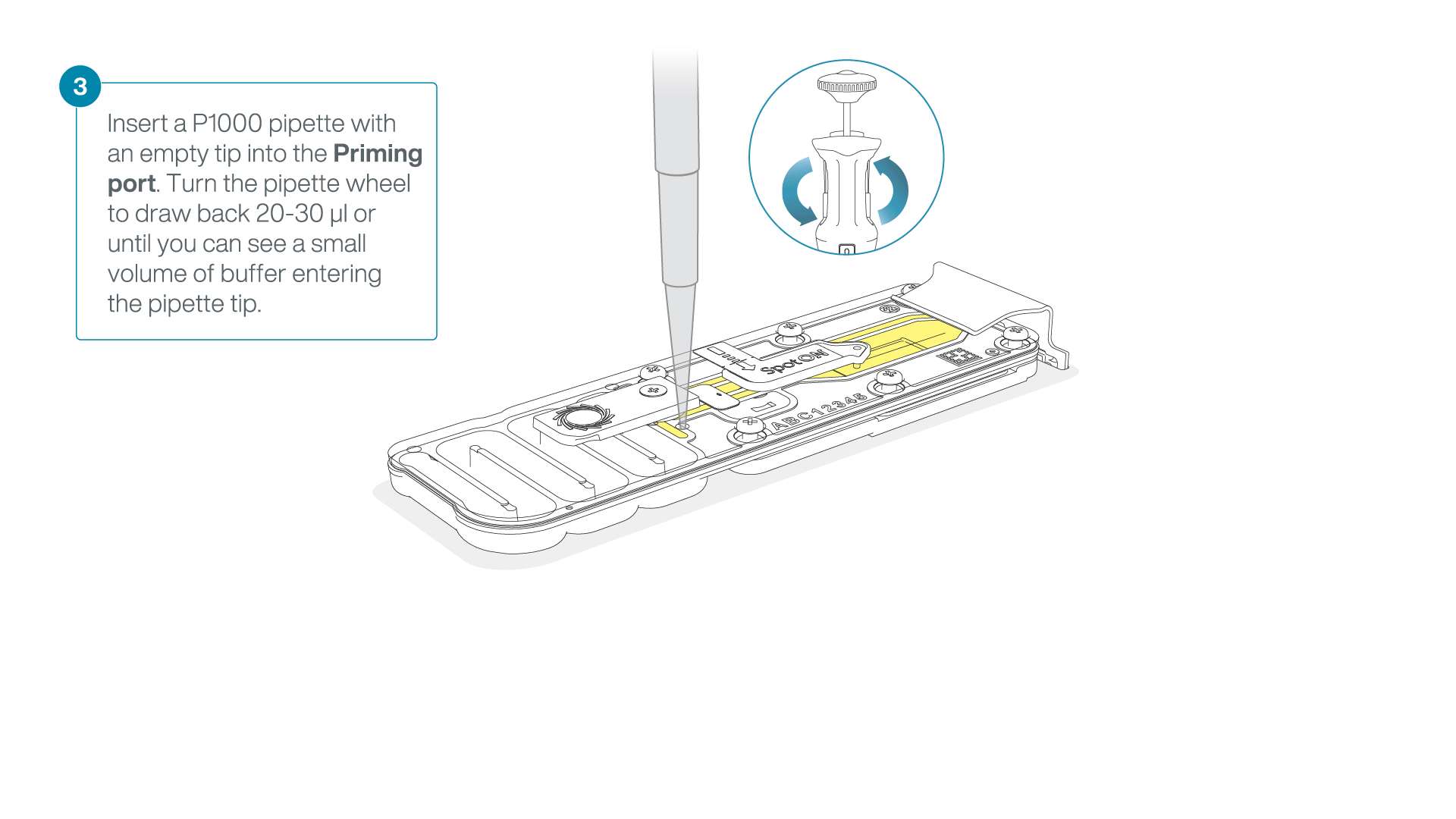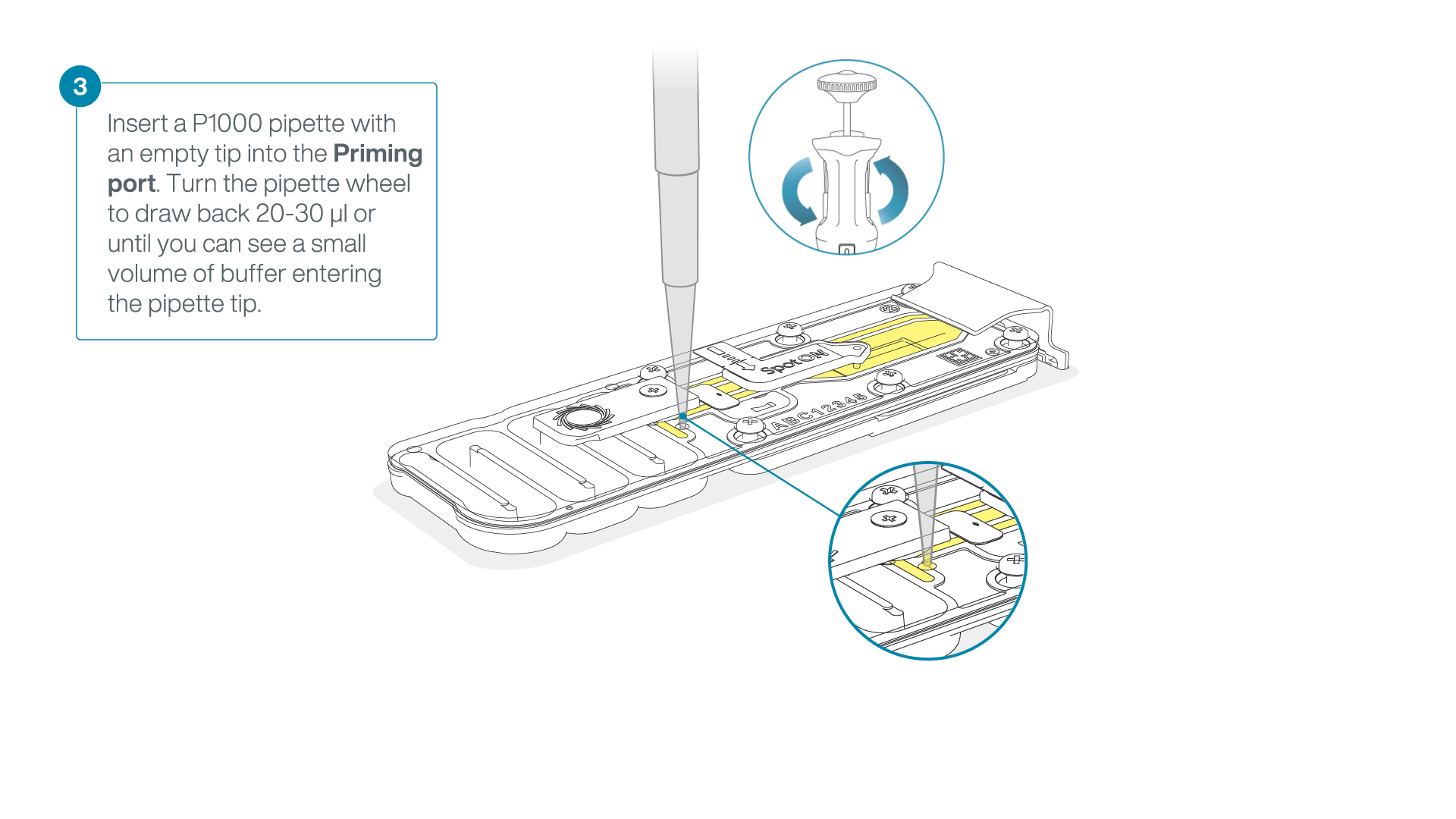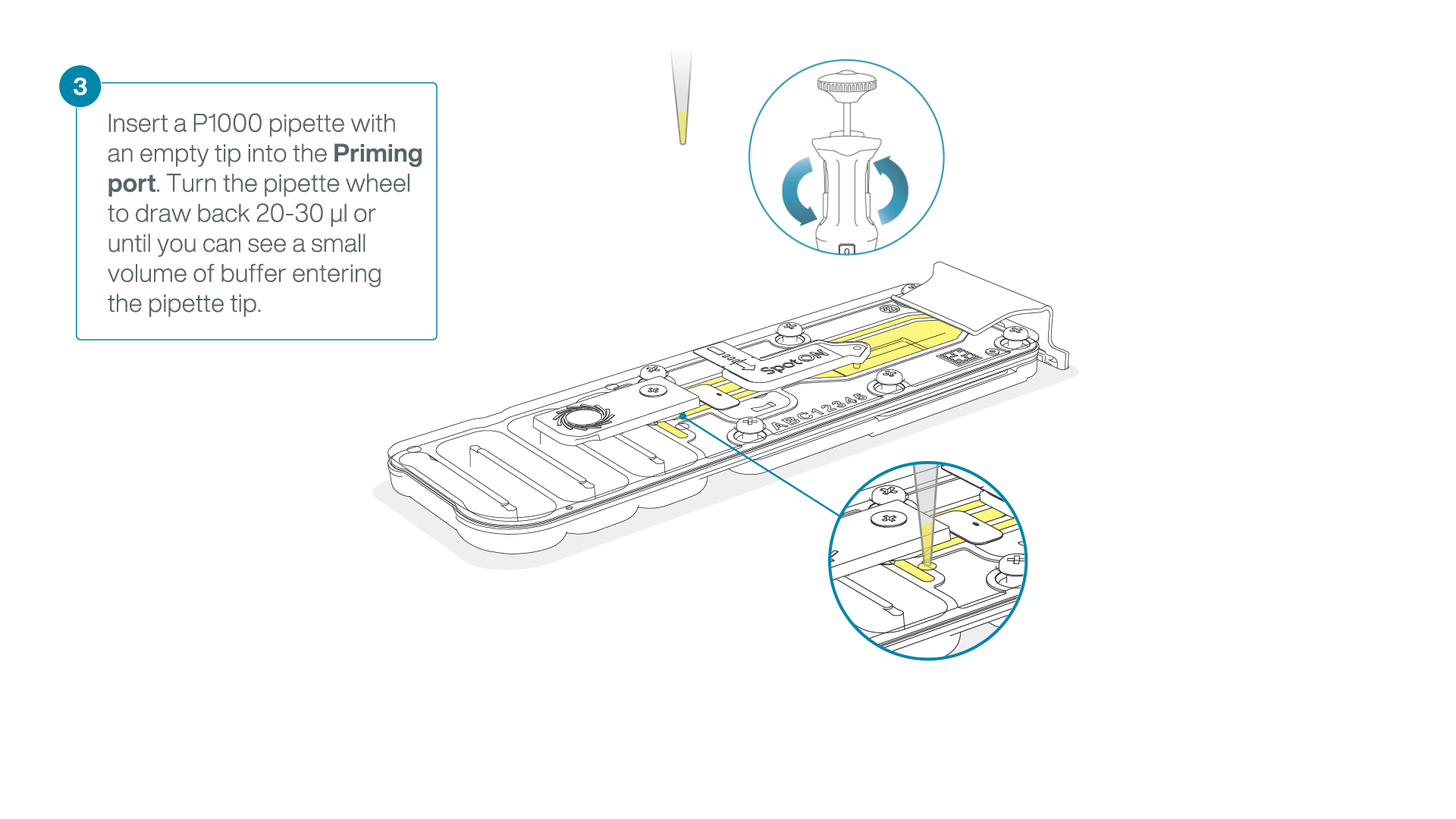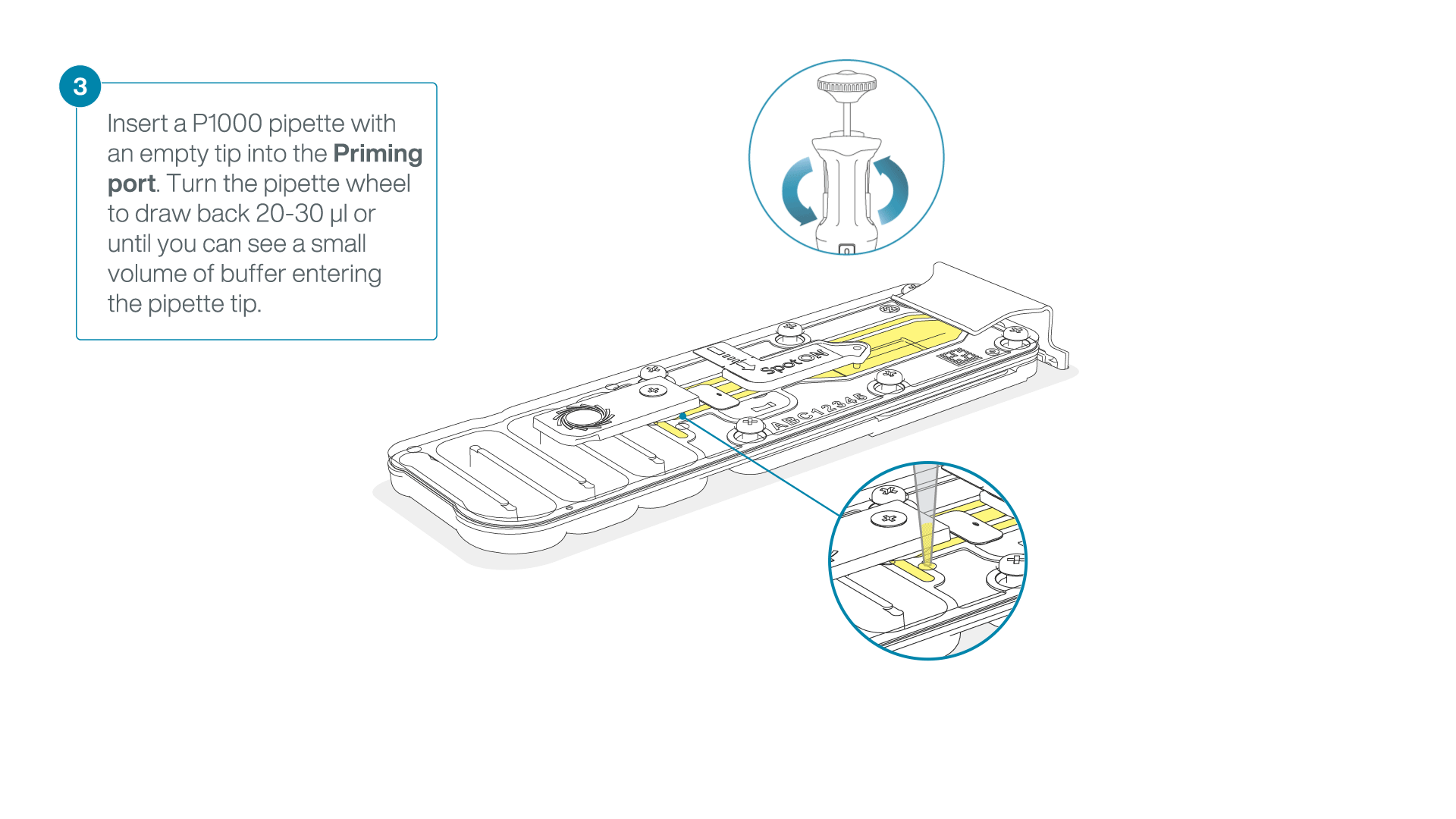
IMPORTANT
Take care when drawing back buffer from the flow cell. Do not remove more than 20-30 µl, and make sure that the array of pores are covered by buffer at all times. Introducing air bubbles into the array can irreversibly damage pores.
Load 400 µl of the prepared Flow Cell Wash Mix into the flow cell priming port, avoiding the introduction of air.
Close the priming port and wait for 30 minutes.
Before removing the waste fluid, ensure that the flow cell priming port cover and SpotON sample port cover are closed, as indicated in the figure below.
Remove all fluid from the waste channel through waste port 1 using a P1000 pipette.
As both the flow cell priming port and SpotON sample port are closed, no fluid should leave the sensor array area.

IMPORTANT
It is vital that the flow cell priming port and SpotON sample port are closed before removing the waste buffer to prevent air from being drawn across the sensor array area, which would lead to a significant loss of sequencing channels.
END OF STEP
Follow one of the two options described in the next steps of the protocol.
- Run a second library on the flow cell straight away
- Store the flow cell for later use
4. To run a second library on a MinION/GridION flow cell straight away
Materials
- Flow Cell Wash Kit (EXP-WSH003)
- Sequencing Auxiliary Vials (EXP-AUX001)
- Flow Cell Priming Kit (EXP-FLP002)
Equipment
- P1000 pipette and tips
- P20 pipette and tips
- Ice bucket with ice
- MinION Flow Cell
TIP
The buffers used in this process are incompatible with conducting a Flow Cell Check prior to loading a subsequent library. However, the first pore scan once a sequencing run has started will report the number of nanopores available.
To run a second library straight away, follow the instructions in the "Priming and loading the flow cell" section of your library preparation protocol with the recommendations below.
Note: For more consistent and better performance after a flushing a flow cell with the Flow Cell Wash Kit, we recommend the following:
- Pipette very slowly when loading priming mix into the flow cell.
- Wait five minutes between priming mix flushes.
- After the five minute pause, close the priming port, ensure the SpotON port is closed and remove the waste from waste port 1. This prevents the nuclease from diffusing through the flow cell. Repeat this step after the second priming mix flush.
As part of this process, the flow cell will need priming using flow cell priming reagents either in your sequencing kit or separately in the Flow Cell Priming Kit.
Once the flow cell has been primed and loaded, either resume the run in MinKNOW or start a new sequencing experiment. Use Join existing experiment when setting up a sequencing run to use the same settings that were used in a previous experiment.
IMPORTANT
When priming a flow cell after a nuclease wash with the Flow Cell Wash Kit, it is vital to wait five minutes between the priming mix flushes and to remove the waste for effective removal of the nuclease.
Reloading a library
Additional buffers are required for reloading a library following the washing of a flow cell. These can be found in the one of the following expansion kits:
Sequencing Auxiliary Vials V14 (EXP-AUX003). This expansion contains vials of Sequencing Buffer (SB), Elution Buffer (EB), Library Solution (LIS), and Library Beads (LIB) with Kit 14 flow cell priming reagents: Flow Cell Flush (FCF) and Flow Cell Tether (FCT). This expansion is only compatible with our Kit 14 chemistry e.g. SQK-LSK114.
Flow Cell Priming Kit (EXP-FLP004). This expansion contains both Kit 14 flow cell priming reagents required: Flow Cell Flush (FCF) and Flow Cell Tether (FCT). This expansion is only compatible with Kit 14 chemistry.
For our previous chemistries:
Sequencing Auxiliary Vials expansion (EXP-AUX001). This expansion contains vials of Elution Buffer (EB), Sequencing Buffer (SQB), and Loading Beads (LB), additional to those found in DNA sequencing kits for our Kit 9 chemistry.
Sequencing Auxiliary Vials expansion (EXP-AUX002). This expansion contains vials of Sequencing Buffer II (SBII), Elution Buffer (EB), Loading Solution (LS), and Loading Beads II (LBII), additional to those found in Kit 10 and 11 chemistry, such as:
- Kit 10 e.g. SQK-LSK110
- Kit 11 e.g. SQK-PCS111
- Flow Cell Priming Kit (EXP-FLO002). This expansion contains both flow cell priming reagents required: Flush Buffer (FB) and Flush Tether (FLT). This expansion is compatible with Kit 9, 10 and 11 chemistry.
TIP
Library storage recommendations
We recommend storing libraries in Eppendorf DNA LoBind tubes at 4°C for short term storage or repeated use, for example, reloading flow cells between washes. For single use and long-term storage of more than 3 months, we recommend storing libraries at -80°C in Eppendorf DNA LoBind tubes. For further information, please refer to the DNA library stability Know-How document.
5. To store the MinION/GridION flow cell for later use
Materials
- Flow Cell Wash Kit (EXP-WSH003)
Optional equipment
- P1000 pipette and tips
- P20 pipette and tips
Thaw one tube of Storage Buffer (S) at room temperature.
Mix contents thoroughly by pipetting and spin down briefly.
Slide the flow cell priming port cover clockwise to open.
After opening the priming port, check for a small air bubble under the cover. Draw back a small volume to remove any bubbles:
- Set a P1000 pipette to 200 µl.
- Insert the tip into the flow cell priming port.
- Turn the wheel until the dial shows 220-230 µl, or until you can see a small volume of buffer/liquid entering the pipette tip.
- Visually check that there is continuous buffer from the flow cell priming port across the sensor array.

Slowly add 500 μl of Storage Buffer (S) through the flow cell priming port.
Close the priming port.
Remove all fluid from the waste channel through waste port 1 using a P1000 pipette.
As both the flow cell priming port and SpotON sample port are closed, no fluid should leave the sensor array area.

IMPORTANT
It is vital that the flow cell priming port and SpotON sample port are closed before removing the waste buffer to prevent air from being drawn across the sensor array area, which would lead to a significant loss of sequencing channels.
The flow cell can now be stored at 4-8°C.
END OF STEP
When you wish to reuse the flow cell, remove the flow cell from storage, and allow it to warm to room temperature for ~5 minutes.
TIP
Library storage recommendations
We recommend storing libraries in Eppendorf DNA LoBind tubes at 4°C for short term storage or repeated use, for example, reloading flow cells between washes. For single use and long-term storage of more than 3 months, we recommend storing libraries at -80°C in Eppendorf DNA LoBind tubes. For further information, please refer to the DNA library stability Know-How document.
