Ligation sequencing gDNA - exome enrichment (SQK-LSK110) (EXS_9118_v110_revI_10Nov2020)
MinION: Protocol
V EXS_9118_v110_revI_10Nov2020
FOR RESEARCH USE ONLY
Contents
Introduction to the protocol
Library preparation
- 4. DNA fragmentation
- 5. End-prep
- 6. Ligation of PCR adapters
- 7. PCR
- 8. Hybridisation
- 9. Pull-down
- 10. Elution and amplification of DNA
- 11. End-prep
- 12. Adapter ligation and clean-up
- 13. Priming and loading the SpotON flow cell
Sequencing and data analysis
Troubleshooting
1. Overview of the protocol
This is a Legacy product
This kit is soon to be discontinued and we recommend all customers to upgrade to the latest chemistry for their relevant kit which is available on the Store. If customers require further support for any ongoing critical experiments using a Legacy product, please contact Customer Support via email: support@nanoporetech.com. For further information on please see the product update page.
Exome sequencing protocol features
Use this protocol if you:
- Are not interested in analysing the entire genome
- Want greater depth of coverage of a specific exonic region
- Want to analyse a large number of target regions
Introduction to exome sequencing
This protocol describes how to carry out exome sequencing using the Ligation Sequencing Kit (SQK-LSK110) and the Agilent SureSelect method.
Steps in the sequencing workflow:
Prepare for your experiment
You will need to:
- Ensure you have your sequencing kit, the correct equipment and third-party reagents
- Download the software for acquiring and analysing your data
- Check your flow cell to ensure it has enough pores for a good sequencing run
Library preparation
You will need to:
- Fragment your DNA
- Ligate PCR adapters to the DNA ends and amplify the fragments
- Hybridise the DNA to probes provided in the Agilent SureSelect Exome kit, and perform a pulldown
- Elute and amplify the pulled-down fragments
- Prepare the DNA ends for adapter attachment
- Attach sequencing adapters supplied in the kit to the DNA ends
- Prime the flow cell, and load your DNA library into the flow cell
Sequencing and analysis
You will need to:
- Start a sequencing run using the MinKNOW software, which will collect raw data from the device and convert it into basecalled reads
- Start the EPI2ME software and select a workflow for Human Exome mapping, or use minimap2 to align the reads to the human reference genome.
We recommend using the standard SureSelect Target Enrichment System protocol from Agilent Technologies with the following differences:
- The DNA in this protocol is sheared with dsDNA Fragmentase rather than in a Covaris g-TUBE
- DNA purification in this protocol is carried out using Agencourt AMPure XP beads instead of QIAquick PCR purification
- DNA End-prep is carried out using the NEBNext Ultra II End Repair/dA-Tailing Module instead of the Agilent method
- Ligation of PCR adapters is carried out using the NEB Blunt/TA Ligase Master Mix instead of the Agilent method
- No desalting of the capture solution is necessary
- LongAMP Taq is used for amplification of the DNA post-hybridisation
This protocol is for the preparation of a single sample. Multiple samples can be prepared in parallel; each sample will require a separate pull-down and will need to be run on a separate flow cell.
Compatibility of this protocol
This protocol should only be used in combination with:
- Ligation Sequencing Kit (SQK-LSK110)
- Control Expansion (EXP-CTL001)
- PCR Expansion (EXP-PCA001)
- FLO-MIN106 (R9.4.1) flow cells
- Flow Cell Wash Kit (EXP-WSH004)
2. Equipment and consumables
材料
- 3 µg high molecular weight human genomic DNA
- Sequence capture kit (e.g. Agilent SureSelect Human All Exon, Cat# 232866)
- Custom primer mix, 10 μM (IDT) - see below for sequences
- Ligation Sequencing Kit (SQK-LSK110)
- PCR 扩展包(EXP-PCA001)
耗材
- NEBNext dsDNA Fragmentase (M0348L)
- NEB Blunt/TA Ligase Master Mix (NEB, cat # M0367)
- NEBNext 快速连接模块(NEB,E6056)
- Cot-1 DNA (ThermoFisher Scientific 15279-011)
- Dynabeads MyOne Streptavidin T1 (ThermoFisher Scientific, 65601)
- Blocking oligo at 1 mM, sequence 5'-AGGTTAAACACCCAAGCAGACGCCGCAATATCAGCACCAACAGAAACAACC-3'
- LongAmp Taq 2X Master Mix (e.g. NEB, cat # M0287)
- 无核酸酶水(如ThermoFisher,AM9937)
- 新制备的 80% 乙醇(用无核酸酶水配制)
- 新制备的70%乙醇(用无核酸酶水配制)
- 0.5 M EDTA,pH 8(Thermo Scientific, R1021)
- 1.5 ml Eppendorf DNA LoBind 离心管
- 0.2 ml 薄壁PCR管
仪器
- Hula混匀仪(低速旋转式混匀仪)
- 适用于1.5ml Eppendorf 离心管的磁力架
- 迷你离心机
- 涡旋混匀仪
- 热循环仪
- 盛有冰的冰桶
- 计时器
- SpeedVac
- P1000 移液枪和枪头
- P200 移液枪和枪头
- P100 移液枪和枪头
- P20 移液枪和枪头
- P10 移液枪和枪头
- P2 移液枪和枪头
可选仪器
- 标准凝胶电泳设备
- Agilent 生物分析仪(或等效仪器)
- Qubit™ 荧光计(或用于质控检测的等效仪器)
- Eppendorf 5424 离心机(或等效器材)
For this protocol, you will need 3 µg high molecular weight human genomic DNA.
Input DNA
How to QC your input DNA
It is important that the input DNA meets the quantity and quality requirements. Using too little or too much DNA, or DNA of poor quality (e.g. highly fragmented or containing RNA or chemical contaminants) can affect your library preparation.
For instructions on how to perform quality control of your DNA sample, please read the Input DNA/RNA QC protocol.
Chemical contaminants
Depending on how the DNA is extracted from the raw sample, certain chemical contaminants may remain in the purified DNA, which can affect library preparation efficiency and sequencing quality. Read more about contaminants on the Contaminants page of the Community.
Custom primer mix sequences
Please order these sequences at 10 μM from IDT:
Forward primer: 5’ CAATTCGGTCTCCAGTGACTTGCCTGTCGCTCTATCTTC 3’ Reverse Primer: 5’ CAATTCGGTCTCCCACTTTTCTGTTGGTGCTGATATTGC 3’
Ligation Sequencing Kit (SQK-LSK110) contents
| Name | Acronym | Cap colour | No. of vials | Fill volume per vial (µl) |
|---|---|---|---|---|
| DNA CS | DCS | Yellow | 1 | 35 |
| Adapter Mix F | AMX-F | Green | 1 | 40 |
| Ligation Buffer | LNB | Clear | 1 | 200 |
| L Fragment Buffer | LFB | White cap, orange stripe on label | 2 | 1,800 |
| S Fragment Buffer | SFB | Grey | 2 | 1,800 |
| Sequencing Buffer II | SBII | Red | 1 | 500 |
| Elution Buffer | EB | Black | 1 | 200 |
| Loading Beads II | LBII | Pink | 1 | 360 |
| Loading Solution | LS | White cap, pink sticker on label | 1 | 360 |
| Flush Buffer | FB | Blue | 6 | 1,170 |
| Flush Tether | FLT | Purple | 1 | 200 |
3. Computer requirements and software
MinION Mk1B IT requirements
Sequencing on a MinION Mk1B requires a high-spec computer or laptop to keep up with the rate of data acquisition. For more information, refer to the MinION Mk1B IT requirements document.
MinION Mk1C IT requirements
The MinION Mk1C contains fully-integrated compute and screen, removing the need for any accessories to generate and analyse nanopore data. For more information refer to the MinION Mk1C IT requirements document.
MinION Mk1D IT requirements
Sequencing on a MinION Mk1D requires a high-spec computer or laptop to keep up with the rate of data acquisition. For more information, refer to the MinION Mk1D IT requirements document.
Software for nanopore sequencing
MinKNOW
The MinKNOW software controls the nanopore sequencing device, collects sequencing data and basecalls in real time. You will be using MinKNOW for every sequencing experiment to sequence, basecall and demultiplex if your samples were barcoded.
For instructions on how to run the MinKNOW software, please refer to the MinKNOW protocol.
EPI2ME (optional)
The EPI2ME cloud-based platform performs further analysis of basecalled data, for example alignment to the Lambda genome, barcoding, or taxonomic classification. You will use the EPI2ME platform only if you would like further analysis of your data post-basecalling.
For instructions on how to create an EPI2ME account and install the EPI2ME Desktop Agent, please refer to this link.
Check your flow cell
We highly recommend that you check the number of pores in your flow cell prior to starting a sequencing experiment. This should be done within 12 weeks of purchasing for MinION/GridION/PromethION or within four weeks of purchasing Flongle Flow Cells. Oxford Nanopore Technologies will replace any flow cell with fewer than the number of pores in the table below, when the result is reported within two days of performing the flow cell check, and when the storage recommendations have been followed. To do the flow cell check, please follow the instructions in the Flow Cell Check document.
| Flow cell | Minimum number of active pores covered by warranty |
|---|---|
| Flongle Flow Cell | 50 |
| MinION/GridION Flow Cell | 800 |
| PromethION Flow Cell | 5000 |
4. DNA fragmentation
材料
- 3 µg high molecular weight human genomic DNA
耗材
- NEBNext dsDNA Fragmentase (M0348L)
- 无核酸酶水(如ThermoFisher,AM9937)
- 新制备的 80% 乙醇(用无核酸酶水配制)
- 0.5 M EDTA,pH 8(Thermo Scientific, R1021)
- 1.5 ml Eppendorf DNA LoBind 离心管
- 0.2 ml 薄壁PCR管
仪器
- 涡旋混匀仪
- Hula混匀仪(低速旋转式混匀仪)
- 适用于1.5ml Eppendorf 离心管的磁力架
- 盛有冰的冰桶
- 迷你离心机
- 热循环仪
可选仪器
- Agilent 生物分析仪(或等效仪器)
- Eppendorf 5424 离心机(或等效器材)
Prepare the DNA in nuclease-free water.
- Transfer 3 μg genomic DNA into a 1.5 ml Eppendorf DNA LoBind tube
- Adjust the volume to 16 μl with nuclease-free water
- Mix thoroughly by flicking the tube to avoid unwanted shearing
- Spin down briefly in a microfuge
In a 0.2 ml thin-walled PCR tube, mix the following:
| Reagent | Volume |
|---|---|
| DNA | 16 µl |
| 10x Fragmentase Reaction Buffer v2 | 2 µl |
| Total | 18 µl |
Vortex the tube for 3 seconds, and spin down.
Add 2 μl dsDNA Fragmentase to the tube.
Incubate the reaction for 28 minutes at 37°C.
Fragmentation timing
Samples may fragment at different rates; 28 mins has shown to be the optimum time for NA12878 DNA extracted with the QIAGEN Genomic-tip. Users may wish to titrate fragmentation times of 25–32 mins if the fragmented yield is low.
Stop the fragmentation reaction by adding 5 μl of 0.5 M EDTA to the tube.
Vortex the tube for 3 seconds, and spin down.
Resuspend the AMPure XP beads by vortexing.
Transfer the sample to a clean 1.5 ml Eppendorf DNA LoBind tube.
Add 15 µl of resuspended AMPure XP beads to the reaction and mix by pipetting.
Incubate on a Hula mixer (rotator mixer) for 10 minutes at room temperature.
Spin down the sample and pellet the beads on a magnet for 5 mins.
Remove and retain the supernatant in a new 1.5 ml Eppendorf DNA LoBind tube.
Note: Do not discard the supernatant.
Add 40 µl of resuspended AMPure XP beads to the reaction and mix by flicking the tube.
Incubate on a Hula mixer (rotator mixer) for 10 minutes at room temperature.
Prepare 500 μl of fresh 80% ethanol in nuclease-free water.
Spin down the sample and pellet the beads on a magnet for 5 minutes. Keep the tube on the magnet until the eluate is clear and colourless, and pipette off the supernatant.
Keep the tube on the magnet and wash the beads with 200 µl of freshly prepared 80% ethanol without disturbing the pellet. Remove the ethanol using a pipette and discard.
Repeat the previous step.
Spin down and place the tube back on the magnet. Pipette off any residual ethanol. Allow to dry for ~30 seconds, but do not dry the pellet to the point of cracking.
Remove the tube from the magnetic rack and resuspend pellet in 21 µl nuclease-free water. Incubate for 2 minutes at room temperature.
Pellet the beads on a magnet until the eluate is clear and colourless.
Remove and retain 21 µl of eluate into a clean 1.5 ml Eppendorf DNA LoBind tube.
Run a 1 μl aliquot on an Agilent Bioanalyzer to determine fragment length.
The fragment length distribution is expected to be similar to the trace below:
Take the fragmented DNA in 20 µl into the end-prep step. However, at this point it is also possible to store the sample at 4°C overnight.
5. End-prep
材料
- Fragmented DNA in 20 µl
耗材
- 0.2 ml 薄壁PCR管
- 1.5 ml Eppendorf DNA LoBind 离心管
- 无核酸酶水(如ThermoFisher,AM9937)
- NEBNext® Ultra II 末端修复/ dA尾添加模块(NEB,E7546)
- Agencourt AMPure XP 磁珠(Beckman Coulter™, A63881)
- 新制备的 80% 乙醇(用无核酸酶水配制)
仪器
- P1000 移液枪和枪头
- P100 移液枪和枪头
- P10 移液枪和枪头
- 热循环仪
- 迷你离心机
- Hula混匀仪(低速旋转式混匀仪)
- 磁力架
- 盛有冰的冰桶
In a 0.2 ml thin-walled PCR tube, mix the following:
Between each addition, pipette mix 10-20 times.
| Reagent | Volume |
|---|---|
| Fragmented DNA | 20 µl |
| Ultra II End-prep reaction buffer | 7 µl |
| Ultra II End-prep enzyme mix | 3 µl |
| Nuclease-free water | 30 µl |
| Total | 60 µl |
Ensure the components are thoroughly mixed by pipetting, and spin down.
Using a thermal cycler, incubate at 20°C for 30 minutes and 65°C for 30 mins.
Resuspend the AMPure XP beads by vortexing.
Transfer the DNA sample to a clean 1.5 ml Eppendorf DNA LoBind tube.
Add 60 µl of resuspended AMPure XP beads to the end-prep reaction and mix by flicking the tube.
Incubate on a Hula mixer (rotator mixer) for 5 minutes at room temperature.
Prepare 500 μl of fresh 80% ethanol in nuclease-free water.
Spin down the sample and pellet on a magnet until supernatant is clear and colourless. Keep the tube on the magnet, and pipette off the supernatant.
Keep the tube on the magnet and wash the beads with 200 µl of freshly prepared 80% ethanol without disturbing the pellet. Remove the ethanol using a pipette and discard.
Repeat the previous step.
Spin down and place the tube back on the magnet. Pipette off any residual ethanol. Allow to dry for ~30 seconds, but do not dry the pellet to the point of cracking.
Remove the tube from the magnetic rack and resuspend pellet in 31 µl nuclease-free water. Incubate for 2 minutes at room temperature.
Pellet the beads on a magnet until the eluate is clear and colourless, for at least 1 minute.
Remove and retain 31 µl of eluate into a clean 1.5 ml Eppendorf DNA LoBind tube.
Quantify 1 µl of end-prepped DNA using a Qubit fluorometer.
Take forward ~300 ng of end-prepped DNA in 30 µl into adapter ligation. However, at this point it is also possible to store the sample at 4°C overnight.
6. Ligation of PCR adapters
材料
- 300 ng end-prepped DNA in 30 µl
- PCR Adapter (PCA)
耗材
- 无核酸酶水(如ThermoFisher,AM9937)
- NEB Blunt/TA Ligase Master Mix (NEB, cat # M0367)
- 新制备的 80% 乙醇(用无核酸酶水配制)
- 1.5 ml Eppendorf DNA LoBind 离心管
仪器
- 适用于1.5ml Eppendorf 离心管的磁力架
- Hula混匀仪(低速旋转式混匀仪)
- Qubit™ 荧光计(或用于质控检测的等效仪器)
- 迷你离心机
- 涡旋混匀仪
Add the reagents in the order given below, mixing by pipetting 10-20 times between each sequential addition:
| Reagent | Volume |
|---|---|
| End-prepped DNA | 30 µl |
| PCR Adapters (PCA) | 20 µl |
| NEB Blunt/TA Ligase Master Mix | 50 µl |
| Total | 100 µl |
Ensure the components are thoroughly mixed by pipetting, and spin down.
Incubate the reaction for 10 minutes at room temperature.
Resuspend the AMPure XP beads by vortexing.
Add 100 µl of resuspended AMPure XP beads to the end-prep reaction and mix by pipetting.
End-prep cleanup demo
Incubate on a Hula mixer (rotator mixer) for 5 minutes at room temperature.
Prepare 500 μl of fresh 80% ethanol in nuclease-free water.
Spin down the sample and pellet on a magnet. Keep the tube on the magnet, and pipette off the supernatant when clear and colourless.
Keep the tube on the magnet and wash the beads with 200 µl of freshly prepared 80% ethanol without disturbing the pellet. Remove the ethanol using a pipette and discard.
Repeat the previous step.
Spin down and place the tube back on the magnet. Pipette off any residual ethanol. Allow to dry for ~30 seconds, but do not dry the pellet to the point of cracking.
Remove the tube from the magnetic rack and resuspend pellet in 49 µl nuclease-free water. Incubate for 2 minutes at room temperature.
Pellet the beads on a magnet until the eluate is clear and colourless.
Remove and retain 49 µl of eluate into a clean 1.5 ml Eppendorf DNA LoBind tube.
Quantify 1 µl of adapted DNA using a Qubit fluorometer.
Take forward 48 µl of the adapted DNA into the PCR reaction. However, at this point it is also possible to store the sample at 4°C overnight.
7. PCR
材料
- Primer Mix (PRM)
耗材
- LongAmp Taq 2X Master Mix (e.g. NEB, cat # M0287)
- 无核酸酶水(如ThermoFisher,AM9937)
- 新制备的 80% 乙醇(用无核酸酶水配制)
- 1.5 ml Eppendorf DNA LoBind 离心管
- 0.2 ml 薄壁PCR管
仪器
- 热循环仪
- 盛有冰的冰桶
- 适用于1.5ml Eppendorf 离心管的磁力架
- Hula混匀仪(低速旋转式混匀仪)
- 涡旋混匀仪
- Qubit™ 荧光计(或用于质控检测的等效仪器)
In a 0.2 ml thin-walled PCR tube mix the following:
| Reagent | Volume |
|---|---|
| LongAmp Taq 2X Master Mix | 50 µl |
| PRM Adapters (10 μM) | 2 µl |
| Template DNA | 48 µl |
| Total | 100 µl |
Amplify using the following cycling conditions:
| Cycle step | Temperature | Time | No. of cycles |
|---|---|---|---|
| Initial denaturation | 95 °C | 3 mins | 1 |
| Denaturation | 98 °C | 20 secs | 6 (b) |
| Annealing | 62 °C (a) | 15 secs (a) | 6 (b) |
| Extension | 65 °C (c) | 3 mins | 6 (b) |
| Final extension | 65 °C | 3 mins | 1 |
| Hold | 4 °C | ∞ |
a. This is specific to the Oxford Nanopore primer and should be maintained
b. Adjust accordingly if input quantities are altered
c. This temperature is determined by the type of polymerase that is being used (given here for LongAmp Taq polymerase)
Resuspend the AMPure XP beads by vortexing.
Add 100 μl of the resuspended AMPure XP beads to the sample, and mix by flicking the tube.
Incubate on a Hula mixer (rotator mixer) for 5 minutes at room temperature.
Prepare 500 μl of fresh 80% ethanol in nuclease-free water.
Spin down the sample and pellet on a magnet. Keep the tube on the magnet, and pipette off the supernatant when clear and colourless.
Keep the tube on the magnet and wash the beads with 200 µl of freshly prepared 80% ethanol without disturbing the pellet. Remove the ethanol using a pipette and discard.
Repeat the previous step.
Spin down and place the tube back on the magnet. Pipette off any residual ethanol. Allow to dry for ~30 seconds, but do not dry the pellet to the point of cracking.
Remove the tube from the magnetic rack and resuspend the pellet in 35 µl nuclease-free water. Incubate for 2 minutes at room temperature.
Pellet the beads on a magnet until the eluate is clear and colourless.
Remove and retain 35 µl of eluate into a clean 1.5 ml Eppendorf DNA LoBind tube.
Quantify 2 µl of amplified DNA using a Qubit fluorometer.
Take forward 300–1000 ng amplified DNA into the hybridisation step. However, at this point it is also possible to store the sample at 4°C overnight.
8. Hybridisation
材料
- Sequence capture kit (e.g. Agilent SureSelect Human All Exon, Cat# 232866)
耗材
- Cot-1 DNA (ThermoFisher Scientific 15279-011)
- 无核酸酶水(如ThermoFisher,AM9937)
- 0.2 ml 薄壁PCR管
- Blocking oligo at 1 mM, sequence 5'-AGGTTAAACACCCAAGCAGACGCCGCAATATCAGCACCAACAGAAACAACC-3'
仪器
- SpeedVac
- 热循环仪
- 盛有冰的冰桶
- 涡旋混匀仪
- 迷你离心机
In a clean 1.5 ml Eppendorf DNA LoBind tube, mix the following:
| Reagent | Volume |
|---|---|
| DNA library | 300–1000 ng |
| Cot-1 DNA | 5 µg |
| Blocking oligo top | 1 µl |
The volume of the reaction can be variable, as the water is evaporated in step 2. After this, the DNA is reconstituted to a set volume.
Evaporate the water in a SpeedVac at 45ºC for approximately 1 hour.
Poke one or more holes in the lid with a narrow gauge needle. You can also break off the cap, cover with parafilm, and poke a hole in the parafilm.
Reconstitute with nuclease-free water to a final volume of 9 µl. Pipette up and down along the sides of the tube for optimal recovery.
Mix thoroughly by vortexing and spin down for 1 minute.
Move the 9 µl DNA library sample to a 0.2 µl thin-walled PCR tube, close the tube and incubate in the thermal cycler using the following program:
| Stage | Temperature | Time |
|---|---|---|
| Step 1 | 95°C | 5 min |
| Step 2 | 65°C | 5 min |
| Step 3 | 65°C | Hold |
You will now need to prepare the Hybridisation Buffer mix and the RNase Block ready to be combined with the Capture Library reagent from the SureSelect kit. This will then be combined with the adapted, amplified DNA sample.
Once the sample tube is in the thermal cycler, mix the reagents in the table below to make the Hybridisation Buffer:
| Reagent | Volume for 1 reaction |
|---|---|
| SureSelect Hyb 1 (orange cap or bottle) | 6.63 µl |
| SureSelect Hyb 2 (red cap) | 0.27 µl |
| SureSelect Hyb 3 (yellow cap or bottle) | 2.65 µl |
| SureSelect Hyb 4 (black cap or bottle) | 3.45 µl |
| Total | 13 µl |
In the event of precipitation, warm the Hybridisation Buffer at 65°C for 5 minutes. Otherwise, keep buffer at room temperature until it is used for the Hybridisation mix.
Dilute the SureSelect RNase Block (purple cap) in nuclease-free water. Keep the mixture on ice.
| RNase block dilution (parts RNase block:water) | Volume of diluted RNase block |
|---|---|
| 25% (1:3) | 2 μl |
Please note that the 1:3 ratio of the RNase Block dilution is different to the SureSelect protocol.
Prepare the Capture Library Hybridisation Mix according to the table below. Only keep the mixture at room temperature until it is added to sample tube.
| Reagent | Volume for 1 reaction |
|---|---|
| Hybridisation buffer mixture | 13 µl |
| 25% RNase Block solution | 2 µl |
| Capture library (red cap) ≥3 Mb | 5 µl |
| Total | 20 µl |
Do not keep solutions containing the Capture Library at room temperature for longer than 10 mins.
Keeping all reagents at 65°C, add 20 µl of the Capture Hybridisation Mix to the tube containing 9 µl adapted and amplified DNA sample.
Mix by pipetting.
Replace the cap on the tube.
The tube must be closed to minimise evaporation and avoid a negative impact on your results.
Incubate the tube for 16–24 hours at 65°C with a heated lid set at 105°C.
Take your sample forward into the next step.
We do not recommend pausing your library prep at this stage by storing your sample at 4°C overnight.
9. Pull-down
材料
- Sequence capture kit (e.g. Agilent SureSelect Human All Exon, Cat# 232866)
耗材
- Dynabeads MyOne Streptavidin T1 (ThermoFisher Scientific, 65601)
- 0.2 ml 薄壁PCR管
- 无核酸酶水(如ThermoFisher,AM9937)
仪器
- 热循环仪
- 涡旋混匀仪
- 磁力架
- 多通道移液枪和枪头
- Plate mixer
- 迷你离心机
- 盛有冰的冰桶
It is important to maintain bead suspensions at 65°C during the washing procedure below to ensure specificity of capture. Make sure that the SureSelect Wash Buffer 2 is pre-warmed to 65°C before use. Do not use a tissue incubator, or other devices with significant temperature fluctuations, for the incubation steps.
The hybrid capture protocol uses the SureSelect Target Enrichment Box 1 reagents (stored at room temperature), as well as Dynabeads MyOne Streptavidin T1 magnetic beads.
Warm the SureSelect Wash Buffer 2 at 65°C.
Resuspend the Dynabeads MyOne Streptavidin T1 magnetic beads by vortexing.
Add 50 µl of the beads to a fresh 1.5 ml Eppendorf DNA LoBind tube.
Add 200 µl of SureSelect Binding Buffer to the beads.
Mix by pipetting.
Place on a magnetic rack, allow beads to pellet and pipette off supernatant.
Repeat steps 4–6 twice more for a total of three washes.
Resuspend the beads in 200 µl of SureSelect Binding Buffer.
Keep the hybridisation tube at 65°C. Using a multichannel pipette, transfer the whole volume (~25–29 µl) of the hybridisation mixture from the 65°C reaction to the tube containing 200 µl of washed streptavidin beads.
Pipette up and down until beads are fully resuspended.
Incubate the tube on a Hula mixer for 30 mins at room temperature. Make sure the sample is mixing in the tube.
Spin down the sample and pellet on a magnet until supernatant is clear and colourless. Keep the tube on the magnet, and pipette off the supernatant.
Resuspend the beads in 200 µl of SureSelect Wash Buffer 1.
Pipette up and down until beads are fully resuspended.
Incubate the reaction for 15 minutes at room temperature.
Spin down the sample and pellet on a magnet until supernatant is clear and colourless. Keep the tube on the magnet, and pipette off the supernatant.
Resuspend the beads in 200 µl of Wash Buffer 2 pre-warmed at 65°C.
Pipette up and down until beads are fully resuspended.
Transfer the sample to a 0.2 ml thin-walled PCR tube.
Incubate the tube for 10 minutes at 65°C in the thermal cycler.
Transfer the sample to a clean 1.5 ml Eppendorf DNA LoBind tube.
Place on a magnetic rack, allow beads to pellet and pipette off supernatant.
Repeat the wash steps 17–22 twice more for a total of three washes. Make sure all of the wash buffer has been removed during the final wash.
Add 96 µl of nuclease-free water to the sample, and pipette up and down to resuspend the beads. Keep the sample on ice.
Captured DNA remains on the streptavidin beads during the post-capture amplification step.
10. Elution and amplification of DNA
材料
- Custom primer mix, 10 μM (IDT) - see below for sequences
耗材
- 0.2 ml 薄壁PCR管
- LongAmp Taq 2X Master Mix (e.g. NEB, cat # M0287)
- 新制备的 80% 乙醇(用无核酸酶水配制)
- 无核酸酶水(如ThermoFisher,AM9937)
- 1.5 ml Eppendorf DNA LoBind 离心管
仪器
- 热循环仪
- 盛有冰的冰桶
- 适用于1.5ml Eppendorf 离心管的磁力架
- Hula混匀仪(低速旋转式混匀仪)
- 涡旋混匀仪
Custom primer mix sequences
Please order these sequences at 10 μM from IDT:
Forward primer: 5’ CAATTCGGTCTCCAGTGACTTGCCTGTCGCTCTATCTTC 3’ Reverse Primer: 5’ CAATTCGGTCTCCCACTTTTCTGTTGGTGCTGATATTGC 3’
Split the sample into 2x 48 µl aliquots, and prepare the following reaction in duplicate.
In a 0.2 ml thin-walled PCR tube, mix the following:
| Reagent | Volume |
|---|---|
| LongAmp Taq 2x Master Mix | 50 µl |
| Custom primer mix | 2 µl |
| Template DNA | 48 µl |
| Total | 100 µl |
Amplify using the following cycling conditions:
| Cycle step | Temperature | Time | No. of cycles |
|---|---|---|---|
| Initial denaturation | 95°C | 3 mins | 1 |
| Denaturation | 98°C | 20 secs | 14 (b) |
| Annealing | 62°C (a) | 15 secs (a) | 14 (b) |
| Extension | 65°C (c) | 3 mins | 14 (b) |
| Final extension | 65°C | 3 mins | 1 |
| Hold | 4°C | ∞ |
a. This is specific to the primer mix and should be maintained
b. Adjust accordingly if input quantities are altered
c. This temperature is determined by the type of polymerase that is being used (given here for LongAmp Taq polymerase)
Place the amplified sample on a magnetic rack. Once the solution is clear, transfer the supernatant into a clean 1.5 ml Eppendorf DNA Lo-Bind tube. The beads can now be discarded.
Resuspend the AMPure XP beads by vortexing.
Add 180 µl of resuspended AMPure XP beads to the reaction and mix by pipetting.
Incubate on a Hula mixer (rotator mixer) for 5 minutes at room temperature.
Prepare sufficient fresh 80% ethanol in nuclease-free water.
Spin down the sample and pellet on a magnet. Keep the tube on the magnet, and pipette off the supernatant when clear and colourless.
Keep the tube on the magnet and wash the beads with 200 µl of freshly prepared 80% ethanol without disturbing the pellet. Remove the ethanol using a pipette and discard.
Repeat the previous step.
Spin down and place the tube back on the magnet. Pipette off any residual ethanol. Allow to dry for ~30 seconds, but do not dry the pellet to the point of cracking.
Remove the tube from the magnetic rack and resuspend pellet in 25 µl nuclease-free water. Incubate for 2 minutes at room temperature.
Pellet the beads on a magnet until the eluate is clear and colourless.
Remove and retain 25 µl of eluate into a clean 1.5 ml Eppendorf DNA LoBind tube. Pool the two samples together to yield 50 µl eluted sample.
Quantify 1 µl of eluted sample using a Qubit fluorometer.
Run a 1 μl aliquot on an Agilent Bioanalyzer to determine fragment length.
The fragment length distribution is expected to be similar to the trace below:
11. End-prep
材料
- 500 ng–1 µg captured DNA in 48 µl
仪器
- 热循环仪
- 适用于1.5ml Eppendorf 离心管的磁力架
- Hula混匀仪(低速旋转式混匀仪)
- 涡旋混匀仪
- 盛有冰的冰桶
可选仪器
- Qubit™ 荧光计(或用于质控检测的等效仪器)
Perform end-repair and dA-tailing as follows:
Mix the following reagents in a 1.5 ml Eppendorf DNA LoBind tube:
| Reagent | Volume |
|---|---|
| DNA | 48 µl |
| Ultra II End-prep reaction buffer | 7 µl |
| Ultra II End-prep enzyme mix | 3 µl |
| Nuclease-free water | 2 µl |
| Total | 60 µl |
Mix gently by flicking the tube, and spin down.
Transfer the sample to a 0.2 ml thin-walled PCR tube.
Using a thermal cycler, incubate at 20°C for 30 minutes and 65°C for 30 mins.
Resuspend the AMPure XP beads by vortexing.
Transfer the sample to a clean 1.5 ml Eppendorf DNA LoBind tube.
Add 60 µl of resuspended AMPure XP beads to the end-prep reaction and mix by pipetting.
Incubate on a Hula mixer (rotator mixer) for 5 minutes at room temperature.
Prepare 500 μl of fresh 70% ethanol in nuclease-free water.
Spin down the sample and pellet on a magnet. Keep the tube on the magnet, and pipette off the supernatant when clear and colourless.
Keep the tube on the magnet and wash the beads with 200 µl of freshly prepared 70% ethanol without disturbing the pellet. Remove the ethanol using a pipette and discard.
Repeat the previous step.
Spin down and place the tube back on the magnet. Pipette off any residual ethanol. Allow to dry for ~30 seconds, but do not dry the pellet to the point of cracking.
Remove the tube from the magnetic rack and resuspend pellet in 31 µl nuclease-free water. Incubate for 2 minutes at room temperature.
Pellet the beads on a magnet until the eluate is clear and colourless.
Remove and retain 31 µl of eluate into a clean 1.5 ml Eppendorf DNA LoBind tube.
Quantify 1 µl of end-prepped DNA using a Qubit fluorometer - recovery aim >300 ng.
Take forward approximately 300 ng of end-prepped DNA in 30 µl into adapter ligation.
12. Adapter ligation and clean-up
材料
- Adapter Mix F (AMX F)
- 连接测序试剂盒内的连接缓冲液(LNB)
- 短片段缓冲液(SFB)
- 洗脱缓冲液(EB)
耗材
- NEBNext 快速连接模块(NEB,E6056)
- 1.5 ml Eppendorf DNA LoBind 离心管
- Agencourt AMPure XP 磁珠(Beckman Coulter™, A63881)
- 无核酸酶水(如ThermoFisher,AM9937)
仪器
- 磁力架
- 迷你离心机
- 涡旋混匀仪
- P1000 移液枪和枪头
- P100 移液枪和枪头
- P20 移液枪和枪头
- P10 移液枪和枪头
Although the recommended third-party ligase is supplied with its own buffer, the ligation efficiency of Adapter Mix (AMX) is higher when using Ligation Buffer supplied within the Ligation Sequencing Kit.
Spin down the Adapter Mix F (AMX-F) and Quick T4 Ligase, and place on ice.
Thaw Ligation Buffer (LNB) at room temperature, spin down and mix by pipetting. Due to viscosity, vortexing this buffer is ineffective. Place on ice immediately after thawing and mixing.
Thaw the Elution Buffer (EB) at room temperature and mix by vortexing. Then spin down and place on ice.
Thaw one tube of Short Fragment Buffer (SFB) at room temperature and mix by vortexing. Then spin down and place on ice.
In a 1.5 ml Eppendorf DNA LoBind tube, mix in the following order:
| Reagent | Volume |
|---|---|
| DNA sample from the previous step | 30 µl |
| Nuclease-free water | 30 µl |
| Ligation Buffer (LNB) | 25 µl |
| NEBNext Quick T4 DNA Ligase | 10 µl |
| Adapter Mix F (AMX-F) | 5 µl |
| Total | 100 µl |
Ensure the components are thoroughly mixed by pipetting, and spin down.
Incubate the reaction for 10 minutes at room temperature.
If you have omitted the AMPure purification step after DNA repair and end-prep, do not incubate the reaction for longer than 10 minutes.
Resuspend the AMPure XP beads by vortexing.
Add 40 µl of resuspended AMPure XP beads to the reaction and mix by flicking the tube.
Incubate on a Hula mixer (rotator mixer) for 5 minutes at room temperature.
Spin down the sample and pellet on a magnet. Keep the tube on the magnet, and pipette off the supernatant when clear and colourless.
Wash the beads by adding 250 μl Short Fragment Buffer (SFB) - do NOT use Long Fragment buffer (LFB). Flick the beads to resuspend, then return the tube to the magnetic rack and allow the beads to pellet. Remove the supernatant using a pipette and discard.
Repeat the previous step.
Spin down and place the tube back on the magnet. Pipette off any residual supernatant. Allow to dry for ~30 seconds, but do not dry the pellet to the point of cracking.
Remove the tube from the magnetic rack and resuspend the pellet in 15 µl Elution Buffer (EB). Spin down and incubate for 10 minutes at room temperature. For high molecular weight DNA, incubating at 37°C can improve the recovery of long fragments.
Pellet the beads on a magnet until the eluate is clear and colourless, for at least 1 minute.
Remove and retain 15 µl of eluate containing the DNA library into a clean 1.5 ml Eppendorf DNA LoBind tube.
Dispose of the pelleted beads
Quantify 1 µl of eluted sample using a Qubit fluorometer.
Use the average fragment size determined at the end of the “Elution and amplification of DNA” step to calculate the molarity of the sample.
Take 50 fmol of library and make up the volume to 12 μl with EB.
The prepared library is used for loading into the flow cell. Store the library on ice or at 4°C until ready to load.
Library storage recommendations
We recommend storing libraries in Eppendorf DNA LoBind tubes at 4°C for short term storage or repeated use, for example, reloading flow cells between washes. For single use and long-term storage of more than 3 months, we recommend storing libraries at -80°C in Eppendorf DNA LoBind tubes. For further information, please refer to the DNA library stability Know-How document.
If quantities allow, the library may be diluted in Elution Buffer (EB) for splitting across multiple flow cells.
Additional buffer for doing this can be found in the Sequencing Auxiliary Vials expansion (EXP-AUX002), available to purchase separately. This expansion also contains additional vials of Sequencing Buffer II (SQII) and Loading Beads II (LBII), required for loading the libraries onto flow cells.
13. Priming and loading the SpotON flow cell
材料
- Flush Buffer (FB)
- Flush Tether (FLT)
- Loading Beads II (LBII)
- Sequencing Buffer II (SBII)
- Loading Solution (LS)
耗材
- 1.5 ml Eppendorf DNA LoBind 离心管
仪器
- MinION device
- SpotON Flow Cell
- P1000 移液枪和枪头
- P100 移液枪和枪头
- P20 移液枪和枪头
Priming and loading a MinION flow cell
We recommend all new users watch the 'Priming and loading your flow cell' video before your first run.
Using the Loading Solution
We recommend using the Loading Beads II (LBII) for loading your library onto the flow cell for most sequencing experiments. However, if you have previously used water to load your library, you must use Loading Solution (LS) instead of water. Note: some customers have noticed that viscous libraries can be loaded more easily when not using Loading Beads II.
Thaw the Sequencing Buffer II (SBII), Loading Beads II (LBII) or Loading Solution (LS, if using), Flush Tether (FLT) and one tube of Flush Buffer (FB) at room temperature before mixing the reagents by vortexing and spin down at room temperature.
To prepare the flow cell priming mix, add 30 µl of thawed and mixed Flush Tether (FLT) directly to the tube of thawed and mixed Flush Buffer (FB), and mix by vortexing at room temperature.
Open the MinION device lid and slide the flow cell under the clip.
Press down firmly on the flow cell to ensure correct thermal and electrical contact.
Complete a flow cell check to assess the number of pores available before loading the library.
This step can be omitted if the flow cell has been checked previously.
See the flow cell check instructions in the MinKNOW protocol for more information.
Slide the flow cell priming port cover clockwise to open the priming port.
Take care when drawing back buffer from the flow cell. Do not remove more than 20-30 µl, and make sure that the array of pores are covered by buffer at all times. Introducing air bubbles into the array can irreversibly damage pores.
After opening the priming port, check for a small air bubble under the cover. Draw back a small volume to remove any bubbles:
- Set a P1000 pipette to 200 µl
- Insert the tip into the priming port
- Turn the wheel until the dial shows 220-230 µl, to draw back 20-30 µl, or until you can see a small volume of buffer entering the pipette tip
Note: Visually check that there is continuous buffer from the priming port across the sensor array.
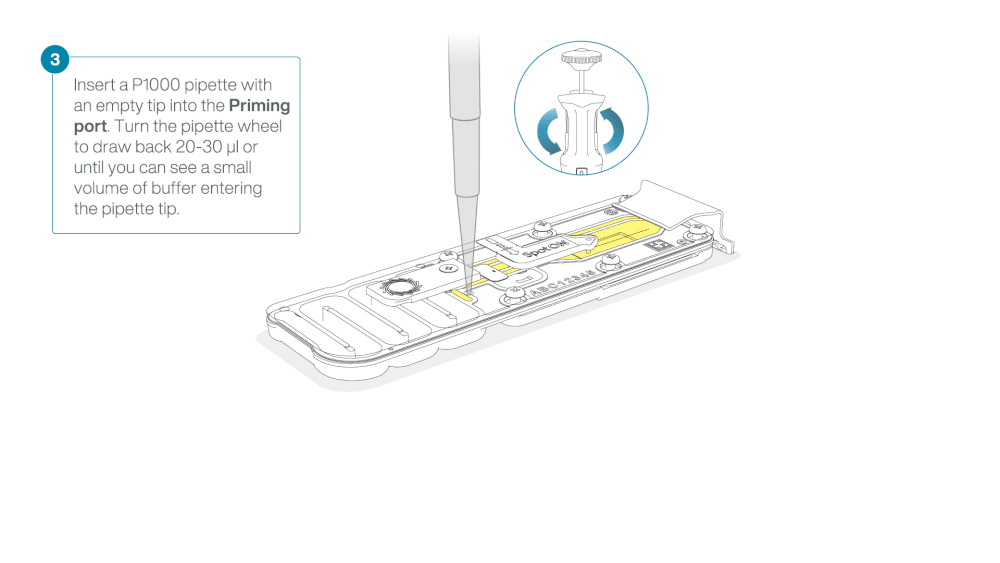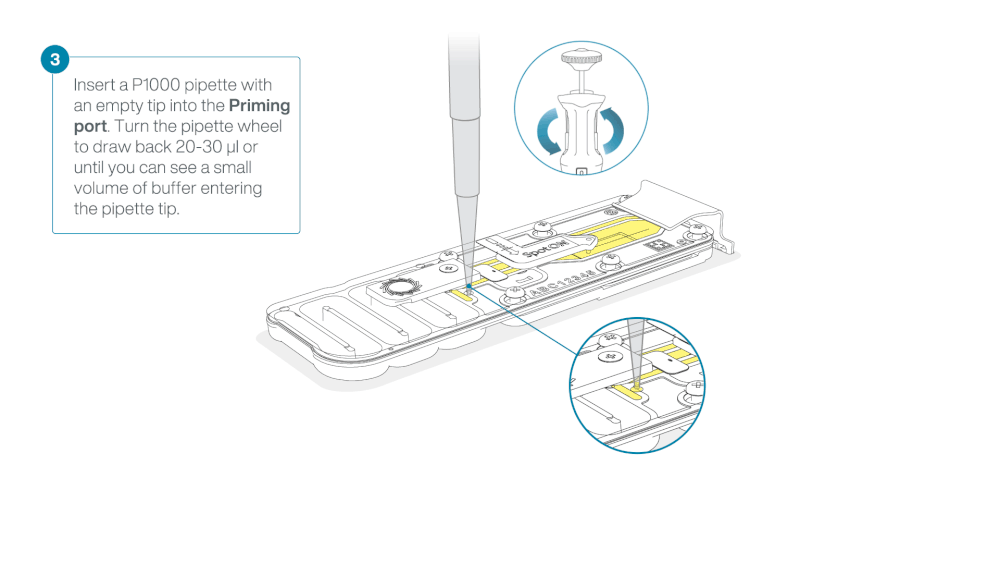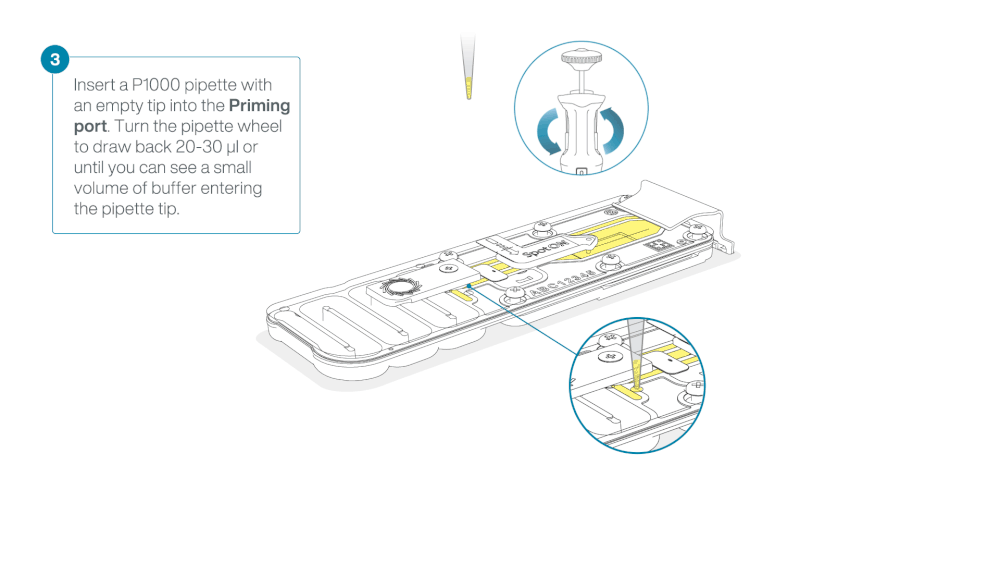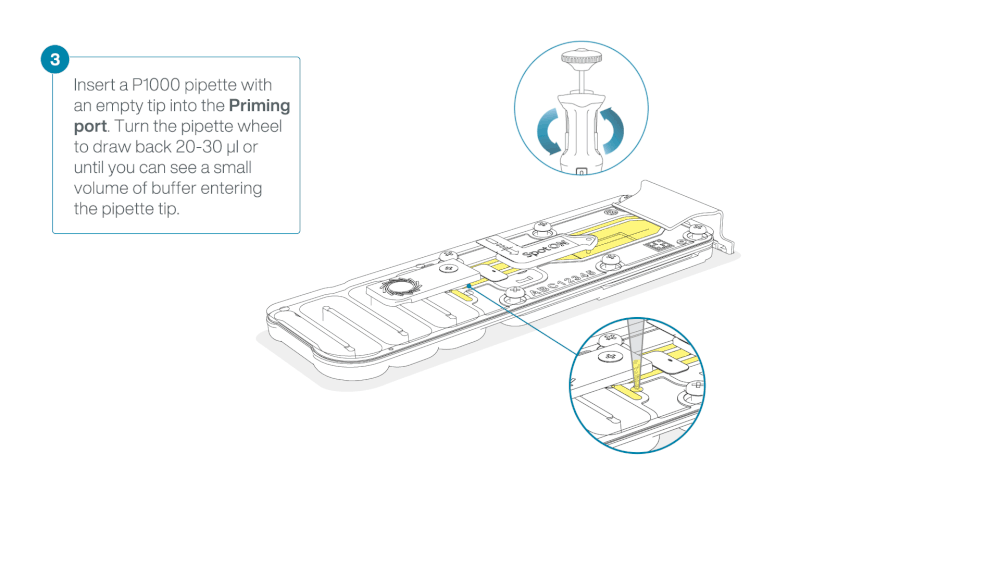
Load 800 µl of the priming mix into the flow cell via the priming port, avoiding the introduction of air bubbles. Wait for five minutes. During this time, prepare the library for loading by following the steps below.

Thoroughly mix the contents of the Loading Beads II (LBII) by pipetting.
The Loading Beads II (LBII) tube contains a suspension of beads. These beads settle very quickly. It is vital that they are mixed immediately before use.
In a new tube, prepare the library for loading as follows:
| Reagent | Volume per flow cell |
|---|---|
| Sequencing Buffer II (SBII) | 37.5 µl |
| Loading Beads II (LBII) mixed immediately before use, or Loading Solution (LS), if using | 25.5 µl |
| DNA library | 12 µl |
| Total | 75 µl |
Note: Load the library onto the flow cell immediately after adding the Sequencing Buffer II (SBII) and Loading Beads II (LBII).
Complete the flow cell priming:
- Gently lift the SpotON sample port cover to make the SpotON sample port accessible.
- Load 200 µl of the priming mix into the flow cell priming port (not the SpotON sample port), avoiding the introduction of air bubbles.

Mix the prepared library gently by pipetting up and down just prior to loading.
Add 75 μl of the prepared library to the flow cell via the SpotON sample port in a dropwise fashion. Ensure each drop flows into the port before adding the next.

Gently replace the SpotON sample port cover, making sure the bung enters the SpotON port, close the priming port and replace the MinION device lid.
14. Data acquisition and basecalling
Overview of nanopore data analysis
For a full overview of nanopore data analysis, which includes options for basecalling and post-basecalling analysis, please refer to the Data Analysis document.
How to start sequencing
The sequencing device control, data acquisition and real-time basecalling are carried out by the MinKNOW software. Please ensure MinKNOW is installed on your computer or device. There are multiple options for how to carry out sequencing:
1. Data acquisition and basecalling in real-time using MinKNOW on a computer
Follow the instructions in the MinKNOW protocol beginning from the "Starting a sequencing run" section until the end of the "Completing a MinKNOW run" section.
2. Data acquisition and basecalling in real-time using the MinION Mk1B/Mk1D device
Follow the instructions in the MinION Mk1B user manual or the MinION Mk1D user manual.
3. Data acquisition and basecalling in real-time using the MinION Mk1C device
Follow the instructions in the MinION Mk1C user manual.
4. Data acquisition and basecalling in real-time using the GridION device
Follow the instructions in the GridION user manual.
5. Data acquisition and basecalling in real-time using the PromethION device
Follow the instructions in the PromethION user manual or the PromethION 2 Solo user manual.
6. Data acquisition using MinKNOW on a computer and basecalling at a later time using MinKNOW
Follow the instructions in the MinKNOW protocol beginning from the "Starting a sequencing run" section until the end of the "Completing a MinKNOW run" section. When setting your experiment parameters, set the Basecalling tab to OFF. After the sequencing experiment has completed, follow the instructions in the Post-run analysis section of the MinKNOW protocol.
15. Downstream analysis
FASTQ Human Exome workflow in EPI2ME
Human Exome is an EPI2ME™ workflow for aligning basecalled reads to a set of reference sequences from Ensembl using the minimap2 aligner, covering the entire human exome. Alignments are performed against full gene sequences, including exons and introns, but currently not against flanking regions. The reference sequences are taken from the feature strand of the most recent human genome assembly, GRCh38.
The experiment report gives details on sequence length, accuracy, quality score and the amount of data generated during the experiment. It is possible to align to a number of human reference genes, and view more details about alignment coverage and accuracy.
For more information, please refer to the Workflows section in the EPI2ME protocol.
Suggested workflow
- Align reads to the human reference genome (GRCh38 or GRCh37) using minimap2. We recommend using -k parameter value 13
- (Optional) Remove reads that do not overlap with pull-down regions
Example data
Expected mean target coverage from one MinION Mk 1B flow cell:
Aligned read length: 
Other data analysis options
1. Bioinformatics tutorials
For more in-depth data analysis, Oxford Nanopore Technologies offers a range of bioinformatics tutorials, which are available in the Bioinformatics resource section of the Community. The tutorials take the user through installing and running pre-built analysis pipelines, which generate a report with the results. The tutorials are aimed at biologists who would like to analyse data without the help of a dedicated bioinformatician, and who are comfortable using the command line.
2. Research analysis tools
Oxford Nanopore Technologies' Research division has created a number of analysis tools, which are available in the Oxford Nanopore GitHub repository. The tools are aimed at advanced users, and contain instructions for how to install and run the software. They are provided as-is, with minimal support.
3. Community-developed analysis tools
If a data analysis method for your research question is not provided in any of the resources above, we recommend the Community-developed data analysis tool library. Numerous members of the Nanopore Community have developed their own tools and pipelines for analysing nanopore sequencing data, most of which are available on GitHub. Please be aware that these tools are not supported by Oxford Nanopore Technologies, and are not guaranteed to be compatible with the latest chemistry/software configuration.
16. Flow cell reuse and returns
材料
- 测序芯片清洗剂盒(EXP-WSH004)
After your sequencing experiment is complete, if you would like to reuse the flow cell, please follow the Flow Cell Wash Kit protocol and store the washed flow cell at +2°C to +8°C.
The Flow Cell Wash Kit protocol is available on the Nanopore Community.
We recommend you to wash the flow cell as soon as possible after you stop the run. However, if this is not possible, leave the flow cell on the device and wash it the next day.
Alternatively, follow the returns procedure to send the flow cell back to Oxford Nanopore.
Instructions for returning flow cells can be found here.
If you encounter issues or have questions about your sequencing experiment, please refer to the Troubleshooting Guide that can be found in the online version of this protocol.
17. Issues during DNA/RNA extraction and library preparation
Below is a list of the most commonly encountered issues, with some suggested causes and solutions.
We also have an FAQ section available on the Nanopore Community Support section.
If you have tried our suggested solutions and the issue still persists, please contact Technical Support via email (support@nanoporetech.com) or via LiveChat in the Nanopore Community.
Low sample quality
| Observation | Possible cause | Comments and actions |
|---|---|---|
| Low DNA purity (Nanodrop reading for DNA OD 260/280 is <1.8 and OD 260/230 is <2.0–2.2) | The DNA extraction method does not provide the required purity | The effects of contaminants are shown in the Contaminants document. Please try an alternative extraction method that does not result in contaminant carryover. Consider performing an additional SPRI clean-up step. |
| Low RNA integrity (RNA integrity number <9.5 RIN, or the rRNA band is shown as a smear on the gel) | The RNA degraded during extraction | Try a different RNA extraction method. For more info on RIN, please see the RNA Integrity Number document. Further information can be found in the DNA/RNA Handling page. |
| RNA has a shorter than expected fragment length | The RNA degraded during extraction | Try a different RNA extraction method. For more info on RIN, please see the RNA Integrity Number document. Further information can be found in the DNA/RNA Handling page. We recommend working in an RNase-free environment, and to keep your lab equipment RNase-free when working with RNA. |
Low DNA recovery after AMPure bead clean-up
| Observation | Possible cause | Comments and actions |
|---|---|---|
| Low recovery | DNA loss due to a lower than intended AMPure beads-to-sample ratio | 1. AMPure beads settle quickly, so ensure they are well resuspended before adding them to the sample. 2. When the AMPure beads-to-sample ratio is lower than 0.4:1, DNA fragments of any size will be lost during the clean-up. |
| Low recovery | DNA fragments are shorter than expected | The lower the AMPure beads-to-sample ratio, the more stringent the selection against short fragments. Please always determine the input DNA length on an agarose gel (or other gel electrophoresis methods) and then calculate the appropriate amount of AMPure beads to use. 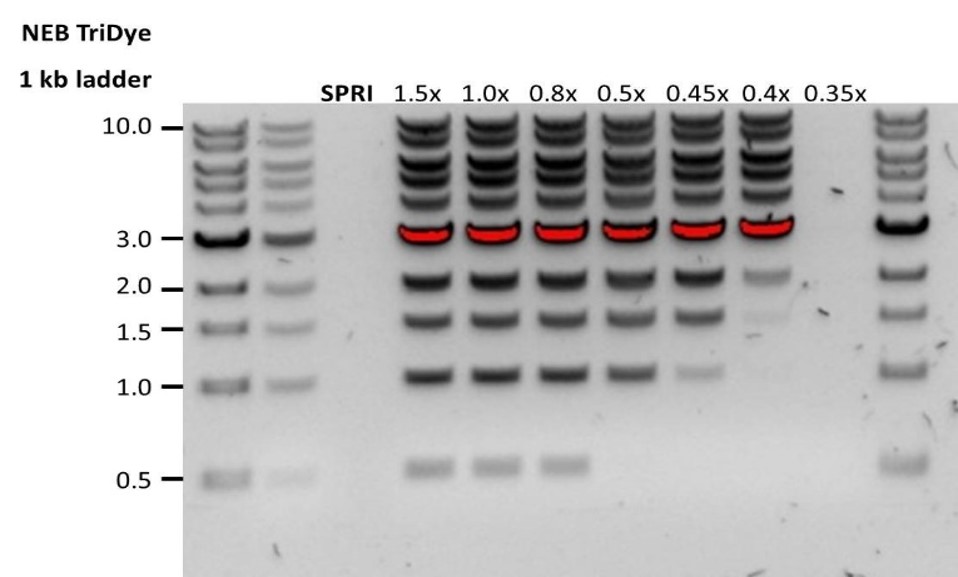 |
| Low recovery after end-prep | The wash step used ethanol <70% | DNA will be eluted from the beads when using ethanol <70%. Make sure to use the correct percentage. |
18. Issues during the sequencing run
Below is a list of the most commonly encountered issues, with some suggested causes and solutions.
We also have an FAQ section available on the Nanopore Community Support section.
If you have tried our suggested solutions and the issue still persists, please contact Technical Support via email (support@nanoporetech.com) or via LiveChat in the Nanopore Community.
Fewer pores at the start of sequencing than after Flow Cell Check
| Observation | Possible cause | Comments and actions |
|---|---|---|
| MinKNOW reported a lower number of pores at the start of sequencing than the number reported by the Flow Cell Check | An air bubble was introduced into the nanopore array | After the Flow Cell Check it is essential to remove any air bubbles near the priming port before priming the flow cell. If not removed, the air bubble can travel to the nanopore array and irreversibly damage the nanopores that have been exposed to air. The best practice to prevent this from happening is demonstrated in this video. |
| MinKNOW reported a lower number of pores at the start of sequencing than the number reported by the Flow Cell Check | The flow cell is not correctly inserted into the device | Stop the sequencing run, remove the flow cell from the sequencing device and insert it again, checking that the flow cell is firmly seated in the device and that it has reached the target temperature. If applicable, try a different position on the device (GridION/PromethION). |
| MinKNOW reported a lower number of pores at the start of sequencing than the number reported by the Flow Cell Check | Contaminations in the library damaged or blocked the pores | The pore count during the Flow Cell Check is performed using the QC DNA molecules present in the flow cell storage buffer. At the start of sequencing, the library itself is used to estimate the number of active pores. Because of this, variability of about 10% in the number of pores is expected. A significantly lower pore count reported at the start of sequencing can be due to contaminants in the library that have damaged the membranes or blocked the pores. Alternative DNA/RNA extraction or purification methods may be needed to improve the purity of the input material. The effects of contaminants are shown in the Contaminants Know-how piece. Please try an alternative extraction method that does not result in contaminant carryover. |
MinKNOW script failed
| Observation | Possible cause | Comments and actions |
|---|---|---|
| MinKNOW shows "Script failed" | Restart the computer and then restart MinKNOW. If the issue persists, please collect the MinKNOW log files and contact Technical Support. If you do not have another sequencing device available, we recommend storing the flow cell and the loaded library at 4°C and contact Technical Support for further storage guidance. |
Pore occupancy below 40%
| Observation | Possible cause | Comments and actions |
|---|---|---|
| Pore occupancy <40% | Not enough library was loaded on the flow cell | Ensure you load the recommended amount of good quality library in the relevant library prep protocol onto your flow cell. Please quantify the library before loading and calculate mols using tools like the Promega Biomath Calculator, choosing "dsDNA: µg to pmol" |
| Pore occupancy close to 0 | The Ligation Sequencing Kit was used, and sequencing adapters did not ligate to the DNA | Make sure to use the NEBNext Quick Ligation Module (E6056) and Oxford Nanopore Technologies Ligation Buffer (LNB, provided in the sequencing kit) at the sequencing adapter ligation step, and use the correct amount of each reagent. A Lambda control library can be prepared to test the integrity of the third-party reagents. |
| Pore occupancy close to 0 | The Ligation Sequencing Kit was used, and ethanol was used instead of LFB or SFB at the wash step after sequencing adapter ligation | Ethanol can denature the motor protein on the sequencing adapters. Make sure the LFB or SFB buffer was used after ligation of sequencing adapters. |
| Pore occupancy close to 0 | No tether on the flow cell | Tethers are adding during flow cell priming (FLT/FCT tube). Make sure FLT/FCT was added to FB/FCF before priming. |
Shorter than expected read length
| Observation | Possible cause | Comments and actions |
|---|---|---|
| Shorter than expected read length | Unwanted fragmentation of DNA sample | Read length reflects input DNA fragment length. Input DNA can be fragmented during extraction and library prep. 1. Please review the Extraction Methods in the Nanopore Community for best practice for extraction. 2. Visualise the input DNA fragment length distribution on an agarose gel before proceeding to the library prep. 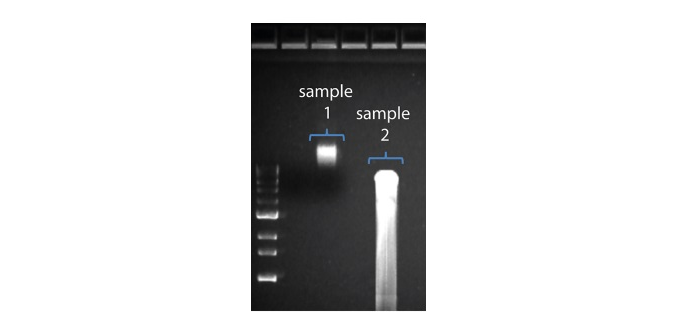 In the image above, Sample 1 is of high molecular weight, whereas Sample 2 has been fragmented. In the image above, Sample 1 is of high molecular weight, whereas Sample 2 has been fragmented.3. During library prep, avoid pipetting and vortexing when mixing reagents. Flicking or inverting the tube is sufficient. |
Large proportion of unavailable pores
| Observation | Possible cause | Comments and actions |
|---|---|---|
Large proportion of unavailable pores (shown as blue in the channels panel and pore activity plot) 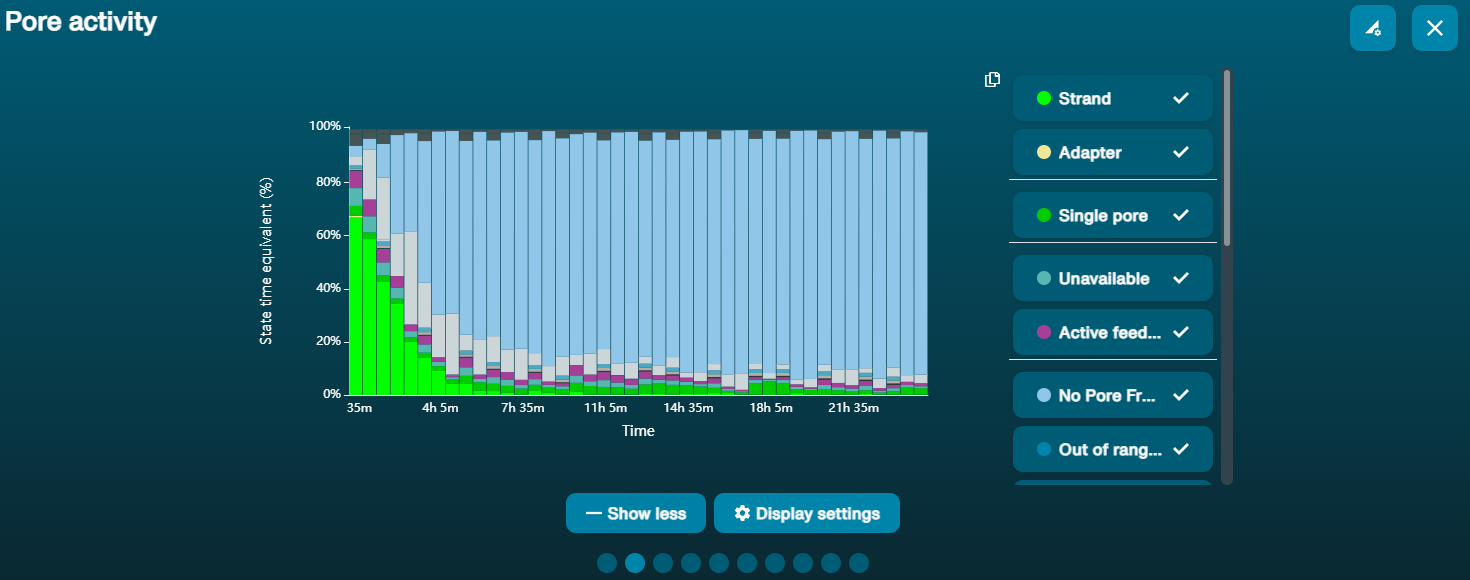 The pore activity plot above shows an increasing proportion of "unavailable" pores over time. The pore activity plot above shows an increasing proportion of "unavailable" pores over time. | Contaminants are present in the sample | Some contaminants can be cleared from the pores by the unblocking function built into MinKNOW. If this is successful, the pore status will change to "sequencing pore". If the portion of unavailable pores stays large or increases: 1. A nuclease flush using the Flow Cell Wash Kit (EXP-WSH004) can be performed, or 2. Run several cycles of PCR to try and dilute any contaminants that may be causing problems. |
Large proportion of inactive pores
| Observation | Possible cause | Comments and actions |
|---|---|---|
| Large proportion of inactive/unavailable pores (shown as light blue in the channels panel and pore activity plot. Pores or membranes are irreversibly damaged) | Air bubbles have been introduced into the flow cell | Air bubbles introduced through flow cell priming and library loading can irreversibly damage the pores. Watch the Priming and loading your flow cell video for best practice |
| Large proportion of inactive/unavailable pores | Certain compounds co-purified with DNA | Known compounds, include polysaccharides, typically associate with plant genomic DNA. 1. Please refer to the Plant leaf DNA extraction method. 2. Clean-up using the QIAGEN PowerClean Pro kit. 3. Perform a whole genome amplification with the original gDNA sample using the QIAGEN REPLI-g kit. |
| Large proportion of inactive/unavailable pores | Contaminants are present in the sample | The effects of contaminants are shown in the Contaminants Know-how piece. Please try an alternative extraction method that does not result in contaminant carryover. |
Reduction in sequencing speed and q-score later into the run
| Observation | Possible cause | Comments and actions |
|---|---|---|
| Reduction in sequencing speed and q-score later into the run | For Kit 9 chemistry (e.g. SQK-LSK109), fast fuel consumption is typically seen when the flow cell is overloaded with library (please see the appropriate protocol for your DNA library to see the recommendation). | Add more fuel to the flow cell by following the instructions in the MinKNOW protocol. In future experiments, load lower amounts of library to the flow cell. |
Temperature fluctuation
| Observation | Possible cause | Comments and actions |
|---|---|---|
| Temperature fluctuation | The flow cell has lost contact with the device | Check that there is a heat pad covering the metal plate on the back of the flow cell. Re-insert the flow cell and press it down to make sure the connector pins are firmly in contact with the device. If the problem persists, please contact Technical Services. |
Failed to reach target temperature
| Observation | Possible cause | Comments and actions |
|---|---|---|
| MinKNOW shows "Failed to reach target temperature" | The instrument was placed in a location that is colder than normal room temperature, or a location with poor ventilation (which leads to the flow cells overheating) | MinKNOW has a default timeframe for the flow cell to reach the target temperature. Once the timeframe is exceeded, an error message will appear and the sequencing experiment will continue. However, sequencing at an incorrect temperature may lead to a decrease in throughput and lower q-scores. Please adjust the location of the sequencing device to ensure that it is placed at room temperature with good ventilation, then re-start the process in MinKNOW. Please refer to this link for more information on MinION temperature control. |
Guppy – no input .fast5 was found or basecalled
| Observation | Possible cause | Comments and actions |
|---|---|---|
| No input .fast5 was found or basecalled | input_path did not point to the .fast5 file location | The --input_path has to be followed by the full file path to the .fast5 files to be basecalled, and the location has to be accessible either locally or remotely through SSH. |
| No input .fast5 was found or basecalled | The .fast5 files were in a subfolder at the input_path location | To allow Guppy to look into subfolders, add the --recursive flag to the command |
Guppy – no Pass or Fail folders were generated after basecalling
| Observation | Possible cause | Comments and actions |
|---|---|---|
| No Pass or Fail folders were generated after basecalling | The --qscore_filtering flag was not included in the command | The --qscore_filtering flag enables filtering of reads into Pass and Fail folders inside the output folder, based on their strand q-score. When performing live basecalling in MinKNOW, a q-score of 7 (corresponding to a basecall accuracy of ~80%) is used to separate reads into Pass and Fail folders. |
Guppy – unusually slow processing on a GPU computer
| Observation | Possible cause | Comments and actions |
|---|---|---|
| Unusually slow processing on a GPU computer | The --device flag wasn't included in the command | The --device flag specifies a GPU device to use for accelerate basecalling. If not included in the command, GPU will not be used. GPUs are counted from zero. An example is --device cuda:0 cuda:1, when 2 GPUs are specified to use by the Guppy command. |







