适用于 16S 与 ITS 的微生物扩增子条形码测序试剂盒(SQK-MAB114.24) (MAB_9229_v114_revB_26Nov2025)
MinION: Protocol
适用于 16S 与 ITS 的微生物扩增子条形码测序试剂盒(SQK-MAB114.24) V MAB_9229_v114_revB_26Nov2025
本实验方案可对多种样本中的 16S rRNA 和 ITS 区域进行扩增及全长测序,分别用于细菌与真菌群落分析。本试剂盒及方法具有以下特点:
- 16S 引物覆盖性广,可提升分类单元的检出能力,同时支持 ITS 引物
- 实现细菌属水平的鉴定
- 支持多达24个不同样本的混样测序
- 流程快捷,相较连接法更加高效、经济
- 采用无片段化工作流程,完整保留扩增子序列
仅供研究使用
本品为早期试用产品
FOR RESEARCH USE ONLY
概览
本实验方案可对多种样本中的 16S rRNA 和 ITS 区域进行扩增及全长测序,分别用于细菌与真菌群落分析。本试剂盒及方法具有以下特点:
- 16S 引物覆盖性广,可提升分类单元的检出能力,同时支持 ITS 引物
- 实现细菌属水平的鉴定
- 支持多达24个不同样本的混样测序
- 流程快捷,相较连接法更加高效、经济
- 采用无片段化工作流程,完整保留扩增子序列
仅供研究使用
本品为早期试用产品
1. 实验方案概览
微生物扩增子条形码测序试剂盒-24 V14 简介
本实验方案详细描述了使用 微生物扩增子条形码测序试剂盒-24 V14(SQK-MAB114.24)为 16S 与 ITS 扩增子添加条形码并建库测序的操作流程及常见问题的解决方法。16S rRNA 基因同时包含高度保守区域(便于设计通用引物并提供系统发育信息)和高度可变区域(不同物种之间存在差异),因此常用于细菌群落分析;而 ITS 区域可用于多种样本类型的真菌群落分析。
微生物扩增子条形码测序试剂盒 24 V14 配备了专门设计的 16S 和 ITS 引物,用于扩增常见的微生物鉴定区域。该靶向测序策略专注于信息量丰富的区域,帮助您准确解析样本的微生物组成,而无需对整个基因组进行全面测序,从而提升实验效率并降低成本。此外,本试剂盒提供 24 种独特条形码,可在一次测序中实现多达 24 个样本的混合测序,同时完整保留全长扩增子信息。
测序完成后,您可以使用 EPI2ME 16S 工作流程(wf-16s) 对样本中的 16S 和 ITS 扩增子进行包括分类在内的下游分析。
测序工作流程:
实验准备
您将需要:
- 提取 gDNA,并根据起始DNA/RNA质控实验指南评估其长度、浓度和纯度。 质量评估步骤对确保实验成功至关重要。
- 确保您已准备好测序试剂盒、正确的仪器以及第三方试剂
- 下载数据收集和分析软件
- 检查您的测序芯片上有足够的活性纳米孔,以确保测序良好运行
文库制备
下表概述了文库制备所需的步骤,包括时间安排和可以中止的节点。
| 实验步骤 | 流程 | 时间 | 中止节点 |
|---|---|---|---|
| 16S 或 ITS PCR 扩增 | 使用试剂盒提供的引物扩增 16S 或 ITS 基因 | 10 分钟 + PCR | 4°C 过夜 |
| 为扩增子添加条形码 | 为多达 24 个扩增子样本添加条形码 | 15 分钟 | |
| 条形码反应终止、样本合并及磁珠纯化 | 终止条形码反应,将带条形码的样本混合,并进行纯化 | 40 分钟 | 若需短期保存或重复使用(如芯片清洗后再次上样),建议置于 4 °C; 若为长期保存且一次性使用,建议置于 –80 °C |
| 快速测序接头连接 | 将快速测序接头连接到 DNA 的末端。 | 5 分钟 | 我们强烈建议您在为文库连接接头后,尽快测序。 |
| 测序芯片预处理及上样 | 对测序芯片进行预处理,然后将 DNA 文库加至芯片中进行测序 | 10 分钟 |
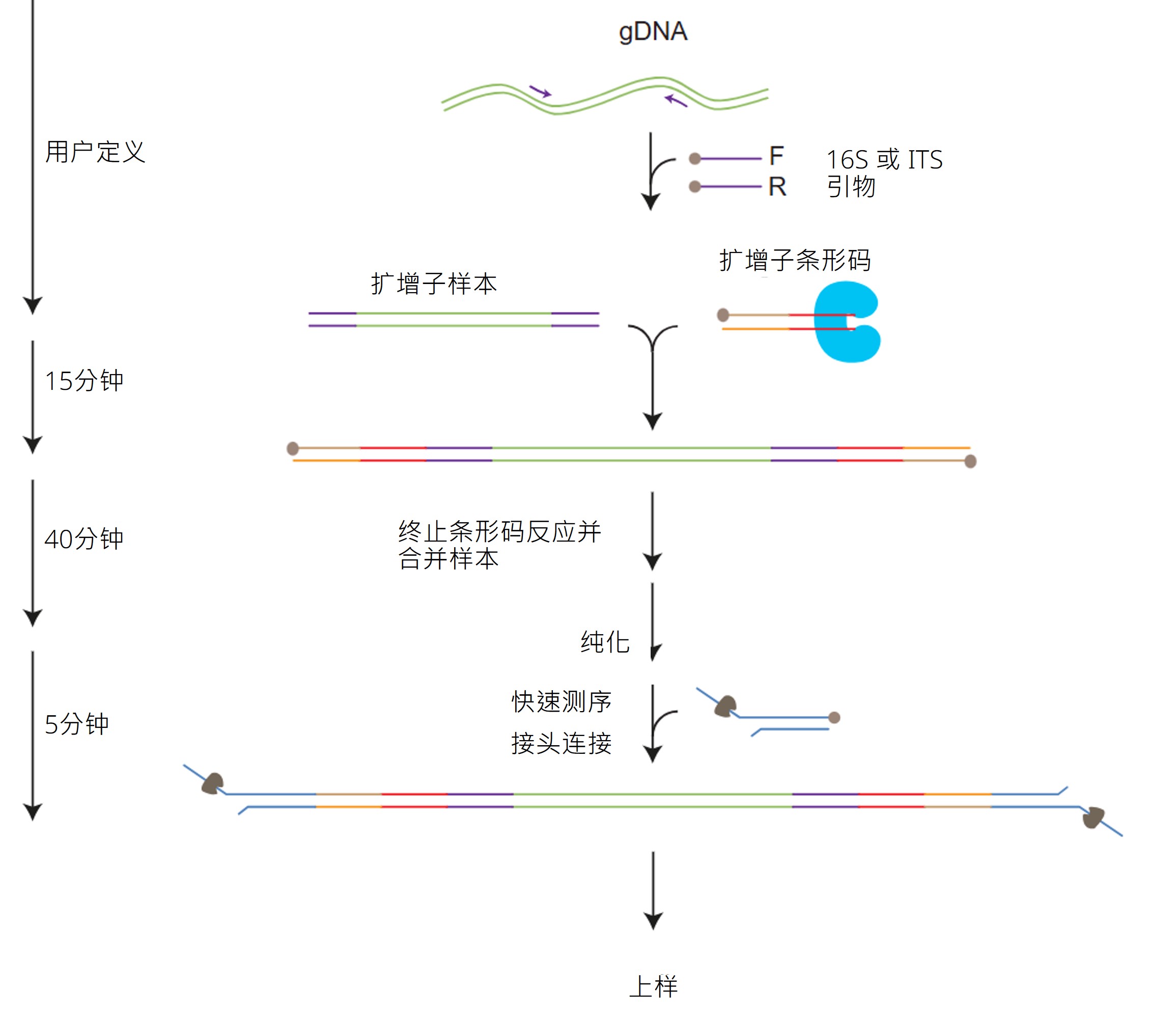
测序和分析
您将需要:
- 使用 MinKNOW 软件开始测序。该软件会通过测序仪收集原始数据,并将其识别成碱基序列。
- 可选步骤:启动 EPI2ME 软件并选择wf-16S 工作流程
实验方案适用性
本实验方案只适用于与以下产品搭配使用:
- 微生物扩增子条形码测序试剂盒-24 V14(SQK-MAB114.24)
- 测序芯片清洗剂盒(EXP-WSH004)
- R10.4.1 测序芯片(FLO-MIN114)
- 快速测序文库接头辅助扩展包 V14 (EXP-RAA114)
- 测序辅助扩展包 V14 (EXP-AUX003)
- 测序芯片预处理试剂盒 (EXP-FLP004)
- MinION Mk1D - MinION Mk1D IT 配置要求文档
- GridION - GridION IT 配置要求文档
2. 仪器及耗材
材料
- 每个样本 10 ng 基因组 DNA
- 微生物扩增子条形码测序试剂盒-24 V14(SQK-MAB114.24)
耗材
- MinION/GridION测序芯片
- 热敏蛋白酶 K(NEB,P8111)
- LongAmp 热启动 Taq 酶 2X 预混液(NEB, M0533)
- Qubit dsDNA HS Assay(双链DNA高灵敏度检测)试剂盒(Invitrogen, Q32851)
- 无核酸酶水(如ThermoFisher,AM9937)
- 1.5 ml Eppendorf DNA LoBind 离心管
- Qubit™ 分析管(Invitrogen, Q32856)
- 0.2 ml 薄壁PCR管
仪器
- MinION 或 GridION 测序仪
- MinION 及GridION 测序芯片遮光片
- Hula混匀仪(低速旋转式混匀仪)
- 迷你离心机
- 涡旋混匀仪
- 适用于1.5ml Eppendorf 离心管的磁力架
- 热循环仪
- P1000 移液枪和枪头
- P200 移液枪和枪头
- P100 移液枪和枪头
- P20 移液枪和枪头
- P10 移液枪和枪头
- P2 移液枪和枪头
- 多通道移液枪和枪头
- 盛有冰的冰桶
- 计时器
- Qubit™ 荧光计(或用于质控检测的等效仪器)
根据本实验指南,您将需要为每个待扩增样本准备 10 ng 高分子量基因组DNA。
第三方试剂
Oxford Nanopore Technologies 推荐您使用本实验指南中列出的所有第三方试剂,并已对其进行验证。我们尚未对其它替代试剂进行测试。
我们建议您按制造商说明准备待用的第三方试剂。
测序芯片质检
我们强烈建议您在开始测序实验前,对测序芯片的活性纳米孔数进行质检。质检需在您收到 MinION/GridION 测序芯片12周内进行。Oxford Nanopore Technologies会对活性孔数量少于以下标准的芯片进行替换*:请您按照测序芯片质检文档中的说明进行芯片质检。
| 测序芯片 | 芯片上的活性孔数确保不少于 |
|---|---|
| MinION/GridION 测序芯片 | 800 |
*(请注意:自收到之日起,芯片须一直贮存于 Oxford Nanopore Technologies 推荐的条件下。且质检结果须在质检后的两日内递交给我们。)
微生物扩增子条形码测序试剂盒-24 V14 内容物
| 名称 | 缩写 | 管盖颜色 | 管数 | 每管溶液体积 (μl) |
|---|---|---|---|---|
| 快速测序文库接头 | RA | 绿色 | 1 | 15 |
| 16S 引物 | 16S | 白色 | 1 | 800 |
| ITS 引物 | ITS | 白色 | 1 | 800 |
| 短片段缓冲液 | SFB | 透明管盖, 白色标签 | 2 | 1800 |
| EDTA | EDTA | 蓝色 | 1 | 700 |
| 接头缓冲液 | ADB | 透明 | 1 | 100 |
| AMPure XP 磁珠 | AXP | 棕色 | 2 | 1200 |
| 洗脱缓冲液 | EB | 黑色 | 1 | 500 |
| 测序缓冲液 | SB | 红色 | 1 | 700 |
| 文库颗粒 | LIB | 粉色 | 1 | 600 |
| 文库溶液 | LIS | 白色管盖,粉色标签 | 1 | 600 |
| 测序芯片冲洗液 | FCF | 透明管盖,浅蓝色标签 | 1 | 8000 |
| 冲洗系绳 UL | FTU | 紫色 | 1 | 65 |
| 扩增子条形码 01-24 | AB24 | - | 两板,每板三套条形码组合 | 每孔 5 μl |
本产品包含由贝克曼库尔特公司(Beckman Coulter, Inc)生产的 AMPure XP 试剂,并可与试剂盒一起于 -20°C 下储存(试剂稳定性将不受损害)。
3. PCR 扩增
材料
- 每个样本 10 ng 基因组 DNA
- 16S 引物
- ITS 引物
耗材
- LongAmp 热启动 Taq 酶 2X 预混液(NEB, M0533)
- Qubit™ dsDNA HS Assay(双链DNA高灵敏度检测)试剂盒(ThermoFisher,Q32851)
- 预冷无核酸酶水(如 ThermoFisher,AM9937)
- 1.5 ml Eppendorf 离心管
- 0.2 ml 薄壁PCR管
仪器
- Hula混匀仪(低速旋转式混匀仪)
- 涡旋混匀仪
- 迷你离心机
- 热循环仪 和/或 恒温干浴器
- P1000 移液枪和枪头
- P200 移液枪和枪头
- P100 移液枪和枪头
- P20 移液枪和枪头
- P10 移液枪和枪头
- P2 移液枪和枪头
- 盛有冰的冰桶
- 计时器
测序芯片质检
我们建议您在开始文库制备之前,对测序芯片的活性纳米孔数量进行质检,以确保其足够支持实验的顺利进行。
详情请参阅 MinKNOW 实验指南中的测序芯片质检说明 。
请将以下试剂解冻,并使用迷你离心机短暂离心后,按照下表说明充分混匀。随后将试剂置于冰上保存备用。
| 试剂 | 1.于室温下解冻 | 2.瞬时离心 | 3.吹打混匀 |
|---|---|---|---|
| 16S 引物 | ✓ | ✓ | ✓ |
| ITS 引物 | ✓ | ✓ | ✓ |
| LongAmp® 热启动 Taq酶 2X 预混液 | ✓ | ✓ | ✓ |
请将 16S 与 ITS 样品分开处理。
请根据样本类型选择正确的引物。
请注意:为获得最佳结果,我们建议您在同一测序芯片上仅测序同一类型的扩增子(如仅 16S 扩增子或仅 ITS 扩增子)。
准备起始DNA样本
- 每个样本取 10 ng 基因组 DNA,分别转移至 0.2 ml 薄壁 PCR 管中;
- 如不足 20 μl,请加入无核酸酶水补足;
- 为避免不必要的打断,请轻弹试管以充分混匀;
- 使用迷你离心机瞬时离心。
在每支含有待测样品的 0.2 ml 薄壁 PCR 管中,配制如下反应体系:
对 16S 样本:
| 试剂 | 体积 |
|---|---|
| 前一步骤所得 10 ng 起始 DNA | 20 μl |
| 16S 引物 | 5 μl |
| LongAmp 热启动 Taq酶 2X 预混液 | 25 μl |
| 总体积 | 50 μl |
注意: 如改变反应体系中的起始材料用量,请适当调整PCR循环数以达到相同产量。
对 ITS 样本:
| 试剂 | 体积 |
|---|---|
| 前一步骤所得 10 ng 起始 DNA | 20 μl |
| ITS 引物 | 5 μl |
| LongAmp 热启动 Taq酶 2X 预混液 | 25 μl |
| 总体积 | 50 μl |
注意: 如改变反应体系中的起始材料用量,请适当调整PCR循环数以达到相同产量。
充分吹打混匀,瞬时离心。
按下表条件进行扩增:
| 步骤 | 温度 | 时间 | 循环数 |
|---|---|---|---|
| 预变性 | 95°C | 1 分钟 | 1 |
| 变性 退火 延伸 | 95°C 55°C 65°C | 20 秒 30 秒 两分钟 | 25 |
| 最后延伸 | 65°C | 5 分钟 | 1 |
| 温度保持 | 4°C | ∞ | - |
扩增结束后,将样品从热循环仪中取出。瞬时离心后分别吹打混匀——请为每个样品更换新的移液枪头。
从每个扩增样本中取 1 µl,采用 Qubit 或等效仪器进行定量质控。
将扩增样品用于后续的条形码连接步骤。
此时所得扩增样品可在 4 °C 下短期保存。
4. 为扩增子连接条形码
材料
- 每个扩增子样品(来自上一步骤)取 10 ng
- 或如使用少于 12 个扩增子条形码,则每个扩增子样品取 20 ng
- 扩增子条形码 01-24(AB01-24)
- 短片段缓冲液(SFB)
- 洗脱缓冲液(EB)
- AMPure XP 磁珠(AXP)
耗材
- 热敏蛋白酶 K(NEB,P8111)
- Qubit™ dsDNA HS Assay(双链DNA高灵敏度检测)试剂盒(ThermoFisher,Q32851)
- 预冷无核酸酶水(如 ThermoFisher,AM9937)
- 1.5 ml Eppendorf 离心管
- 0.2 ml 薄壁PCR管
仪器
- Hula混匀仪(低速旋转式混匀仪)
- 涡旋混匀仪
- 迷你离心机
- 热循环仪 和/或 恒温干浴器
- P1000 移液枪和枪头
- P200 移液枪和枪头
- P100 移液枪和枪头
- P20 移液枪和枪头
- P10 移液枪和枪头
- P2 移液枪和枪头
- 盛有冰的冰桶
- 计时器
使用较少数量的条形码时,对样本起始量的最低要求
当扩增子条形码数目不足 12 个时,为保证最佳测序效果,建议每个样本使用 20 ng 扩增子 DNA。
请将以下试剂解冻,并使用迷你离心机短暂离心后,按照下表说明充分混匀。随后将试剂置于冰上保存备用。
| 试剂 | 1.于室温下解冻 | 2.瞬时离心 | 3.吹打混匀 |
|---|---|---|---|
| 扩增子条形码 01-24(AB01-24) | ✓ | ✓ | ✓ |
| EDTA(EDTA) | ✓ | ✓ | ✓ |
| 短片段缓冲液(SFB) | ✓ | ✓ | ✓ |
| AMPure XP 磁珠(AXP) | ✓ | ✓ | 临使用前,吹打或涡旋混匀 |
| 洗脱缓冲液(EB) | ✓ | ✓ | ✓ |
| 热敏蛋白酶 K | ✓ | ✓ | ✓ |
条形码板孔仅限一次性使用。使用前请确认所选孔密封完好;一旦刺穿或开启,不得再次使用。
使用无核酸酶水配制扩增子 DNA 样本。
- 将每个扩增子 DNA 样品(来自之前步骤)分别转移至单独的 0.2 ml 薄壁 PCR 管中:
| 样本数 ≥ 12 | 样本数 ≤ 11 |
|---|---|
| 每管加入 10 ng 扩增子 DNA 样本 | 每管加入 20 ng 扩增子 DNA 样本 |
- 如不足8 μl,请加入无核酸酶水补足
- 为避免不必要的打断,请轻弹试管以充分混匀
- 使用迷你离心机瞬时离心
为计划上样于同一测序芯片的各个样本选择不同的扩增子条形码。
一个实验中最多可包含24个样本。
请注意:每个样本应仅对应一种条形码。为获得最佳结果,我们建议您在同一测序芯片上仅测序同一类型的扩增子(如仅 16S 扩增子或仅 ITS 扩增子)。
在每支含有待测扩增子样品的 0.2 ml 薄壁 PCR 管中,配制如下反应体系:
| 试剂 | 体积 |
|---|---|
| 之前步骤所得的 16S 或 ITS 扩增子 DNA | 8 μl |
| EDTA | 1 μl |
| 扩增子条形码(AB01-24) | 1 μl |
| 总体积 | 10 μl |
充分吹打混匀,瞬时离心。
将反应在热循环仪中孵育:65℃ 10 分钟,后接 80℃ 两分钟。
从热循环仪内取出样本,并瞬时离心。
向每管反应中加入1 μl的热敏蛋白酶K,然后吹打混匀。
将反应在热循环仪中孵育:37℃ 15分钟,后接 55℃ 10分钟。
从热循环仪内取出样本,并瞬时离心。
将各带条形码的 DNA 样本合并至一支洁净的1.5 ml Eppendorf DNA LoBind离心管中。
| 每个样本的体积 | 6个样本 | 12个样本 | 24个样本 | |
|---|---|---|---|---|
| 总体积 | ~11 μl | 66 μl | 132 μl | 264 μl |
涡旋振荡以重悬AMPure XP磁珠(AXP)。
将重悬后的 AMPure XP 磁珠(AXP)按 0.7 倍体积比例加入含条形码的混合样本,吹打混匀:
| . | 每个样本的体积 | 6个样本 | 12个样本 | 24个样本 |
|---|---|---|---|---|
| AMPure XP 磁珠体积 (AXP) | 7.7 μl | 46 μl | 93 μl | 185 μl |
注意: 表格中列出了示例体积,供您参考。请依据条形码样本混合液的总体积,调整 AMPure XP 磁珠(AXP)的加入量,确保磁珠与样本的体积比为 0.7。
将离心管置于Hula混匀仪(低速旋转式混匀仪)上室温孵育5分钟。
将样品瞬时离心后置于磁力架上,待磁珠与液相分离,且液相澄清无色。保持离心管在磁力架上不动,用移液枪吸去清液。
加入 250 μl短片段缓冲液(SFB)洗涤磁珠。轻弹离心管将磁珠混匀后,将离心管瞬时离心,再放回磁力架,静置待磁珠和液相分离。保持试管在磁力架上不动,用移液枪吸去清液。
重复上述步骤。
将离心管瞬时离心后置于磁力架上。用移液枪吸走残留的上清液。让磁珠在空气中干燥约30秒,但不要干至表面开裂。
将离心管从磁力架上移开。将磁珠重悬于 15µl 洗脱缓冲液中(EB)。瞬时离心,然后在室温下孵育5分钟。
将离心管静置于磁力架上至少1分钟,直到磁珠和液相分离,且洗脱液澄清无色。
将15µl洗脱液转移至一支新的1.5ml Eppendorf DNA LoBind管中。
- 将含有 DNA 文库 的洗脱液转移至新的 1.5 ml Eppendorf DNA LoBind 管中
- 将磁珠丢弃
取1µl洗脱样品,用Qubit定量。
请注意:若文库浓度过低,可能源于纯化步骤的损失。请确保最终文库量充足,以满足测序芯片的上样需求。
扩增子条形码步骤完成后,样本 DNA 的回收率通常应不低于起始投入量的 50%。
将 11 µl 洗脱样品转至洁净的 1.5 ml Eppendorf DNA LoBind 离心管中。
洗脱后的带条形码 DNA 文库可直接用于后续的快速测序接头连接步骤。
在此阶段,该文库可在 4 °C 条件下短期保存;若需长期保存,则请置于 –80 °C。
5. 快速测序文库接头连接
材料
- DNA 文库洗脱液
- 快速测序文库接头(RA)
- 接头缓冲液(ADB)
耗材
- 1.5 ml Eppendorf 离心管
仪器
- 涡旋混匀仪
- 迷你离心机
- P1000 移液枪和枪头
- P200 移液枪和枪头
- P100 移液枪和枪头
- P20 移液枪和枪头
- P10 移液枪和枪头
- P2 移液枪和枪头
- 盛有冰的冰桶
- 计时器
请将以下试剂解冻,并使用迷你离心机短暂离心后,按照下表说明充分混匀。随后将试剂置于冰上保存备用。
| 试剂 | 1.于室温下解冻 | 2.瞬时离心 | 3.吹打混匀 |
|---|---|---|---|
| 快速测序文库接头(RA) | ✓ | ✓ | ✓ |
| 接头缓冲液(ADB) | ✓ | ✓ | ✓ |
在一支新的 1.5 ml Eppendorf DNA LoBind 管中,按以下比例稀释快速测序文库接头(RA),并吹打混匀:
| 试剂 | 体积 |
|---|---|
| 快速测序文库接头(RA) | 1.5 μl |
| 接头缓冲液(ADB) | 3.5 μl |
| 总体积 | 5 μl |
向带有条形码的 DNA 中加入 1 µl 稀释后的快速测序文库接头(RA)。
轻弹离心管以充分混合,并瞬时离心。
室温下孵育5分钟。
构建好的文库即可用于测序芯片上样。在上样前,请将文库置于冰上保存。
6. MinION 及 GridION 测序芯片的预处理及上样
材料
- 测序芯片冲洗液(FCF)
- 冲洗系绳 UL(FTU)
- 文库颗粒(LIB)
- 测序缓冲液(SB)
耗材
- MinION/GridION测序芯片
- 1.5 ml Eppendorf DNA LoBind 离心管
仪器
- MinION 或 GridION 测序仪
- MinION 及GridION 测序芯片遮光片
- P1000 移液枪和枪头
- P100 移液枪和枪头
- P20 移液枪和枪头
- P10 移液枪和枪头
- 涡旋混匀仪
请注意:本试剂盒仅兼容 R10.4.1 测序芯片(FLO-MIN114)。
从冰箱中取出测序芯片,在室温下放置 20 分钟,以便在预处理和上样时更清晰地观察到传感器阵列。
测序芯片的预处理及上样
我们建议所有新用户在首次运行测序芯片前,观看视频测序芯片的预处理及上样。
于室温下解冻测序缓冲液(SB)、文库颗粒(LB)、冲洗系绳 UL(FTU)和测序芯片冲洗液(FCF)。完全解冻后,涡旋振荡混匀。然后瞬时离心,置于冰上。
按下表所示,混合测序芯片冲洗液 (FCF) 与冲洗系绳 UL(FTU) ,制备测序芯片的预处理液。室温下吹打混匀。
请根据待上样测序芯片的数量,另拿一支适当体积的离心管,按下表制备测序芯片预处理液。
| 试剂 | 体积(每张芯片) |
|---|---|
| 测序芯片冲洗液 (FCF) | 1197 µl |
| 冲洗系绳 UL(FTU) | 3 µl |
| 总体积 | 1200 µl |
打开 MinION 或 GridION 测序仪的盖子,将测序芯片插入金属固定夹的下方。用力向下按压预处理孔孔盖处,以确保正确的热、电接触。


为文库上样前,完成测序芯片质检,查看可用孔数目。
如此前已对测序芯片进行过质检,则此步骤可省略。
详情请参阅 MinKNOW 实验指南中的测序芯片质检说明 。
顺时针转动测序芯片的预处理孔孔盖,使预处理孔显露出来。

小心地从测序芯片中反旋吸出缓冲液。请勿吸出超过 20-30 µl的缓冲液,并确保芯片上的纳米孔阵列一直有缓冲液覆盖。将气泡引入阵列会对纳米孔造成不可逆转地损害。
将预处理孔打开后,检查孔周围是否有小气泡。请按照以下方法,从孔中排出少量液体以清除气泡:
1.将 P1000 移液枪转至 200µl 刻度。
2.将枪头垂直插入预处理孔中。
3.反向转动移液枪量程调节转纽,直至移液枪刻度在 220-230 µl之间,或直至您看到有少量缓冲液进入移液枪枪头。
注意: 肉眼检查,确保从预处理孔到传感器阵列的缓冲液连续且无气泡。

通过预处理孔向芯片中加入 800µl 预处理液,避免引入气泡。等待5分钟。在此期间,请按照以下步骤准备用于上样的文库。

将含有文库颗粒的LIB管用移液枪吹打混匀。
LIB管内的文库颗粒分散于悬浮液中。由于颗粒沉降速度非常快,因此请在混匀颗粒后立即使用。
在一支新的1.5ml Eppendorf DNA LoBind离心管内,将所有试剂按以下顺序混合:
| 试剂 | 每张测序芯片的上样体积 |
|---|---|
| 测序缓冲液(SB) | 37.5 µl |
| 文库颗粒 (LIB),临用前混匀;或文库溶液 (LIS) | 25.5 µl |
| DNA 文库 | 12 µl |
| 总体积 | 75 µl |
完成测序芯片的预处理:
- 轻轻地翻起SpotON上样孔盖,使SpotON上样孔显露出来。

- 通过预处理孔(而 非 SpotON加样孔)向芯片中加入200µl预处理液,避免引入气泡。

临上样前,用移液枪轻轻吹打混匀制备好的文库。
通过 SpotON 加样孔向芯片中逐滴加入 75µl 制备好的文库。确保液滴流入孔内后,再加下一滴。

轻轻合上 SpotON 加样孔孔盖,确保塞头塞入加样孔内。合上预处理孔盖。


为获得最佳测序产出,在文库样本上样后,请立即在测序芯片上安装遮光片。
我们建议在清洗芯片并重新上样时,将遮光片保留在测序芯片上。一旦文库从测序芯片中吸出,即可取下遮光片。
按下述步骤安装测序芯片遮光片:
1.小心将遮光片的前沿(平端)与金属固定夹的边沿对齐。 注意: 请勿将遮光片强行压到固定夹下方。
2.将遮光片轻轻盖在测序芯片上。遮光片的 SpotON 加样孔孔盖缺口应与芯片上的 SpotON 加样孔孔盖接合,遮盖住整个测序芯片的前部。

MinION测序芯片的遮光片并非固定在测序芯片上,因此当为芯片加装遮光片后,请小心操作。
合上测序设备上盖,在 MinKNOW 上设置测序实验。
当测序芯片插入 MinION Mk1D 测序仪时,仪器的盖子会遮盖在芯片上方,但盖子与芯片之间会留有空隙。此属正常现象,不影响设备性能。
请参阅此常见问题解答 ,了解有关测序仪上盖的更多信息。

7. 数据采集和碱基识别
如何开始测序
在完成测序芯片的加样后,您即可在MinKNOW中启动测序实验。MinKNOW 软件负责仪器控制、数据采集以及实时碱基识别。有关设置和使用 MinKNOW 的详细信息,请参阅MinKNOW 实验指南。
您可以通过多种方式使用并设置MinKNOW:
- 在直接或远程连接到测序设备的计算机上。
- 直接在 GridION 或 PromethION 24/48 测序设备上。
有关在测序设备上使用 MinKNOW 的更多信息,请参阅相应设备的用户手册:
在MinKNOW中启动测序:
1.在 "开始 "(Start)页面上,选择 开始测序 (Start Sequencing)。
2.输入实验详情:例如实验名称,测序芯片位置及样本ID。
3.在"试剂盒"页面上,选择 所用的建库试剂盒 。
4.在“实验配置”(Run Configuration)页面设置测序与输出参数,或保持默认值。
请注意: 如果在设置实验参数时关闭了碱基识别,您可在实验结束后,在 MinKNOW 中运行线下碱基识别。详情请参阅MinKNOW实验指南。
5.单击 "参数确认" 页面上的 开始 启动测序。
测序后数据分析
当于MinKNOW上完成测序后,您可按照“测序芯片的重复利用及回收”一节中的说明重复使用或返还测序芯片。
完成测序和碱基识别后,即可进行数据分析。有关碱基识别和后续分析选项的详细信息,请参阅数据分析文档。
在下游分析部分,我们将概述更多用于数据分析的选项。
8. 测序芯片的重复利用及回收
材料
- 测序芯片清洗剂盒(EXP-WSH004)
完成测序实验后,如您希望再次使用测序芯片,请按照“测序芯片清洗试剂盒实验指南”进行操作,并将清洗后的芯片置于 +2 至 +8℃ 保存。
您可在纳米孔社区获取测序芯片清洗试剂盒实验指南。
或者,请按照回收程序将测序芯片返还至Oxford Nanopore。
您可在 此处找到回收测序芯片的说明。
如果您遇到问题或对测序实验有疑问,请参阅本实验指南在线版本中的“疑难解答指南”一节。
9. 下游分析
下游分析
我们建议使用 EPI2ME 进行下游分析。EPI2ME 为一款桌面软件,可运行 Nextflow 工作流程,帮助用户高效开展生物信息学分析。平台内的分析流程由长读长测序领域的专家精心开发维护。
点击相应链接,获取有关 EPI2ME 工作流程的更多信息和快速入门指南。
16S 工作流程(wf-16s) 基于 Nextflow,结合 wf-metagenomics 的功能,用于识别靶向扩增子测序中序列片段的来源。该流程可通过 kraken2 或 minimap2 两种模式运行来判定序列片段来源。
请点击此处,查看有关 EPI2ME 16S 工作流程(wf-16s) 的详细信息。
安装说明请参考此处。
此外,您还可以选择其他方法对经过碱基识别的数据进行进一步分析,包括:
1.科研分析工具
Oxford Nanopore Technologies 的研发部门开发了许多分析工具,您可在 Oxford Nanopore 的 GitHub 资料库中找到。这些工具面向有一定经验的用户,并包含如何安装和运行软件的说明。工具是以源代码的形式提供的,因此我们仅提供有限的技术支持。
2.纳米孔社区用户开发的分析工具
如上述资源未能提供满足您研究需求的数据分析方法,请前往资源中心,查找适用的生物信息学工具。该工具库汇总了各种第三方用户开发、且在Github上开源的、针对纳米孔数据的生信分析工具。请注意,Oxford Nanopore Technologies不为这些工具提供支持,也不能保证它们与测序所用的最新的化学试剂/软件配置兼容。
10. DNA 提取和文库制备过程中可能出现的问题
以下表格列出了提取和文库制备过程中的常见问题,以及可能的原因和解决方法。
我们还在 Nanopore 社区的Support板块提供了常见问题解答(FAQ)。
如果以下方案仍无法解决您的问题,请通过电邮(support@nanoporetech.com)或 纳米孔社区的在线支持(LiveChat)联系我们。
低质量样本
经AMPure磁珠纯化后的DNA回收率低
| 现象 | 可能原因 | 措施及备注 |
|---|---|---|
| 低回收率 | AMPure磁珠量与样品量的比例低于预期,导致DNA因未被捕获而丢失 | 1.AMPure 磁珠沉降速度较快,因此在将磁珠加入样品前,请务必充分重悬混匀。 2.当AMPure磁珠量与样品量的比值低于0.4:1时,所有的DNA片段都会在纯化过程中丢失。 |
| 低回收率 | DNA片段短于预期 | AMPure磁珠量与样品量的比值越低,针对短片段的筛选就越严格。每次实验时,请先使用琼脂糖凝胶(或其他凝胶电泳方法)确定起始DNA的长度,据此计算出合适的AMPure磁珠用量。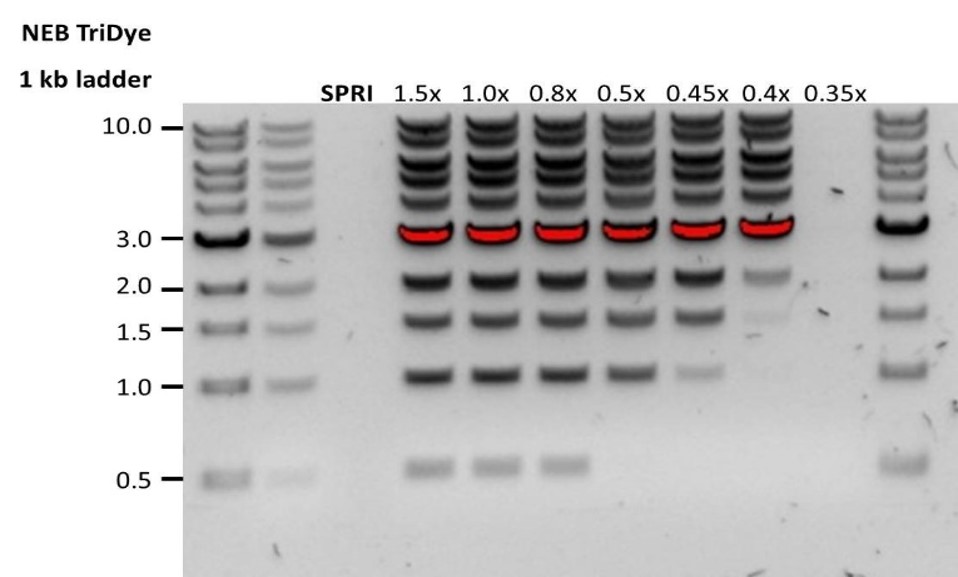 |
| 末端修复后的DNA回收率低 | 清洗步骤所用乙醇的浓度低于80% | 当乙醇浓度低于80%时,DNA会从磁珠上洗脱下来。请确保使用正确浓度的乙醇。 |
11. 使用快速测序试剂盒时,测序过程中可能出现的问题
以下表格列出了提取和文库制备过程中的常见问题,以及可能的原因和解决方法。
我们还在 Nanopore 社区的Support板块提供了常见问题解答(FAQ)。
如果以下方案仍无法解决您的问题,请通过电邮(support@nanoporetech.com)或 纳米孔社区的在线支持(LiveChat)联系我们。
MinKNOW Mux 扫描在测序起始时报告的活性孔数少于芯片质检时报告的活性孔数
| 现象 | 可能原因 | 措施及备注 |
|---|---|---|
| MinKNOW Mux 扫描在测序起始时报告的活性孔数少于芯片质检时报告的活性孔数 | 纳米孔阵列中引入了气泡 | 在对通过质控的芯片进行预处理之前,请务必排出预处理孔附近的气泡。否则,气泡会进入纳米孔阵列对其造成不可逆转地损害。视频中演示了避免引入气泡的最佳操作方法。 |
| MinKNOW Mux 扫描在测序起始时报告的活性孔数少于芯片质检时报告的活性孔数 | 测序芯片没有正确插入测序仪 | 停止测序,将芯片从测序仪中取出,再重新插入测序仪内。请确保测序芯片牢固嵌入测序仪中,并已到达目标温度。如用户使用的是GridION/PromethION测序仪,也可尝试将芯片插入仪器的其它芯片槽进行测序。 |
| MinKNOW Mux 扫描在测序起始时报告的活性孔数少于芯片质检时报告的活性孔数 | 文库中残留的污染物对纳米孔造成损害或堵塞 | 在测序芯片质检阶段,我们用芯片储存缓冲液中的质控DNA分子来评估活性纳米孔的数量。而在测序开始时,我们使用DNA文库本身来评估活性纳米孔的数量。因此,活性纳米孔的数量在这两次评估中会有约10%的浮动。如测序开始时报告的孔数明显降低,则可能是由于文库中的污染物对膜结构造成了损坏或将纳米孔堵塞。用户可能需要使用其它的DNA/RNA提取或纯化方法,以提高起始核酸的纯度。您可在 污染物专题技术文档中查看污染物对测序实验的影响。请尝试其它不会导致污染物残留的 提取方法。 |
MinKNOW脚本失败
| 现象 | 可能原因 | 措施及备注 |
|---|---|---|
| MinKNOW显示 "Script failed”(脚本失败) | 重启计算机及MinKNOW。如问题仍未得到解决,请收集 MinKNOW日志文件并联系我们的技术支持。如您没有其他可用的测序设备,我们建议您先将装有文库的测序芯片置于4°C 储存,并联系我们的技术支持团队获取进一步储存上的建议。 |
纳米孔利用率低于40%
| 现象 | 可能原因 | 措施及备注 |
|---|---|---|
| 纳米孔利用率<40% | 测序芯片中的文库量不够 | 建议向 MinION/GridION 测序芯片中加入 10–50 fmol 的优质文库。请在上样前对文库进行定量,并使用Promega Biomath Calculator等工具中的“dsDNA:µg to pmol”功能来计算 DNA 分子的摩尔量。 |
| 纳米孔利用率接近0 | 使用快速 PCR 条形码测序试剂盒-24,但测序接头并未与 DNA 成功连接 | 请务必严格遵循实验方案,使用准确的体积并保持正确的孵育温度。您可通过制备Lambda对照文库来检验试剂的可用性。 |
| 纳米孔利用率接近0 | 测序芯片中无系绳 | 系绳(FCT 管)随测序芯片预处理液加至芯片。请确保在制备预处理液时,将测序芯片系绳(FCT)加入测序芯片冲洗液(FCF)中。 |
读长短于预期
| 现象 | 可能原因 | 措施及备注 |
|---|---|---|
| 读长短于预期 | DNA样本降解 | 读长反映了起始DNA片段的长度。起始DNA在提取和文库制备过程中均有可能被打断。 1.请查阅纳米孔社区中的 提取方法 以获得最佳DNA提取方案. 2.在进行文库制备之前,请先跑电泳,查看起始DNA片段的长度分布。 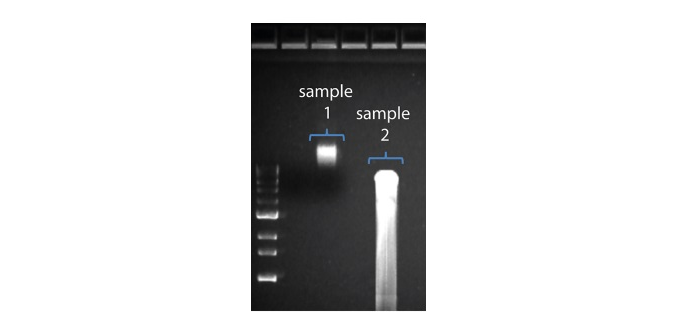 在上图中,样本1为高分子量DNA,而样本2为降解样本。 在上图中,样本1为高分子量DNA,而样本2为降解样本。3.在制备文库的过程中,请避免使用吹打或/和涡旋振荡的方式来混合试剂。轻弹或上下颠倒离心管即可。 |
大量纳米孔处于不可用状态
| 现象 | 可能原因 | 措施及备注 |
|---|---|---|
大量纳米孔处于不可用状态 (在通道面板和纳米孔活动状态图上以蓝色表示) 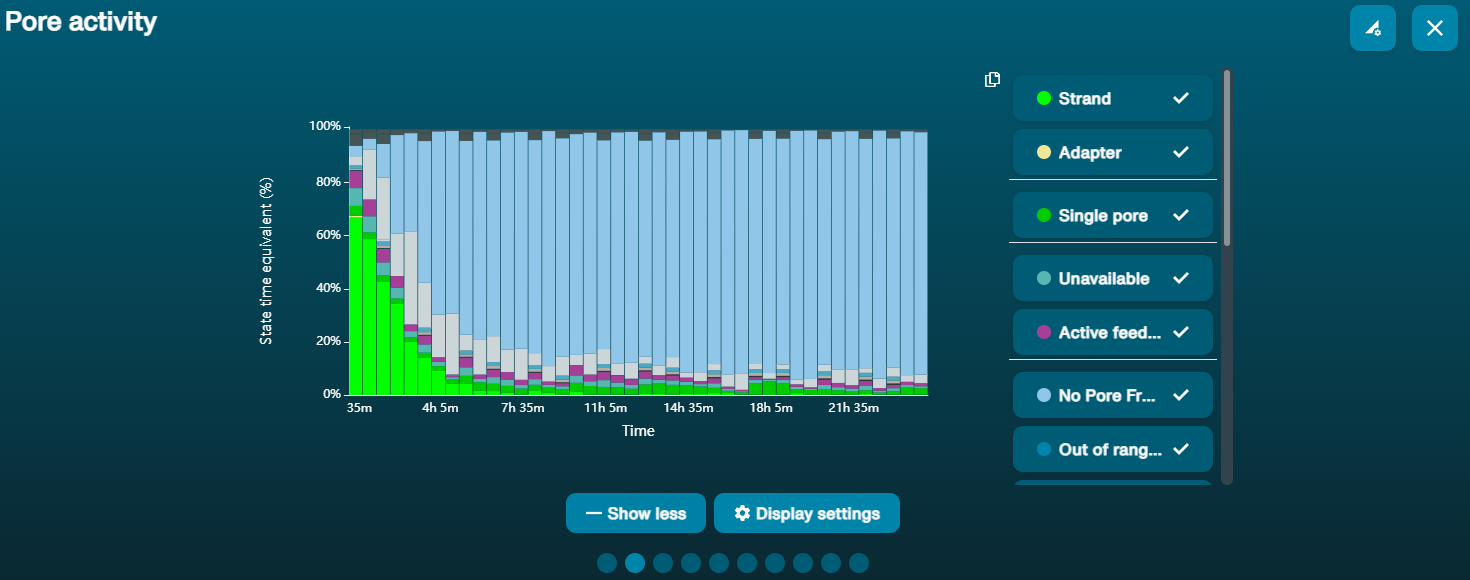 上方的纳米孔活动状态图显示:状态为不可用的纳米孔的比例随着测序进程而不断增加。 上方的纳米孔活动状态图显示:状态为不可用的纳米孔的比例随着测序进程而不断增加。 | 样本中含有污染物 | 使用MinKNOW中的“Unblocking”(疏通)功能,可对一些污染物进行清除。如疏通成功,纳米孔的状态会变为"测序孔"(sequencing pore)。若疏通后,状态为不可用的纳米孔的比例仍然很高甚至增加: 1.用户可使用 测序芯片冲洗试剂盒 (EXP-WSH004)进行核酸酶冲洗 操作,或 2.使用PCR扩增目标片段,以稀释可能导致问题的污染物。 |
大量纳米孔处于“失活”(Inactive)状态
| 现象 | 可能原因 | 措施及备注 |
|---|---|---|
| 大量纳米孔处于失活状态(在通道面板和纳米孔活动状态图上以浅蓝色表示。膜结构或纳米孔遭受不可逆转地损伤 | 测序芯片中引入了气泡 | 芯片预处理和文库上样过程中引入的气泡会对纳米孔带来不可逆转地损害。请观看 测序芯片的预处理及上样 视频了解最佳操作方法 |
| 大量纳米孔处于失活/不可用状态 | 存在与DNA共纯化的化合物 | 已知的化合物包括通常与植物基因组 DNA 相关的多糖等。请参考植物叶片DNA提取方法. 2.使用 QIAGEN PowerClean Pro 试剂盒进行纯化。 3.利用 QIAGEN REPLI-g 试剂盒对原始 gDNA 样本进行全基因组扩增。 |
| 大量纳米孔处于失活/不可用状态 | 样本中含有污染物 | 您可在Contaminants 中查看污染物对测序实验的影响。请尝试其它不会导致污染物残留的提取方法。 |
温度波动
| 现象 | 可能原因 | 措施及备注 |
|---|---|---|
| 温度波动 | 测序芯片和仪器接触不良 | 检查芯片背面的金属板是否有热垫覆盖。重新插入测序芯片,用力向下按压,以确保芯片的连接器引脚与测序仪牢固接触。如问题仍未得到解决,请联系我们的技术支持。 |
未能达到目标温度
| 现象 | 可能原因 | 措施及备注 |
|---|---|---|
| MinKNOW显示“未能达到目标温度” | 测序仪所处环境低于标准室温,或通风不良(以致芯片过热) | MinKNOW会限定测序芯片达到目标温度的时间。当超过限定时间后,系统会显示出错信息,但测序实验仍会继续。值得注意的是,在错误温度下测序可能会导致通量和数据质量(Q值)的降低。请调整测序仪的摆放位置,确保将其置于室温下、通风良好的环境中,再在MinKNOW中重启进程。有关 MinION 温度控制的更多信息,请点击 此链接查看。 |
12. 引物设计、PCR 与测序注意事项
16S 及 ITS rRNA 序列
点击以下文档链接,下载完整的 16S 与 ITS 引物序列:
本试剂盒所采用的引物方案符合行业标准,分别覆盖完整的 16S rRNA 基因和 ITS rRNA 基因。但由于引物结合区域在不同物种间并非完全保守,因而无法保证对所有物种均能成功扩增。
建议您开展对照实验,以确认所研究的目标物种是否能够借助本试剂盒所提供的引物实现扩增。如需进一步支持,请联系support@nanoporetech.com。
ITS 引物设计
本试剂盒所提供的 ITS 引物符合行业标准。
请注意:ITS 引物可能形成二聚体,在 PCR 扩增过程中生成非目标片段,并在后续的文库制备中被保留下来。由此,测序数据中可能出现少量较长或质量较低的序列片段,例如:
串联序列(concatemer reads)
双链序列(2D reads),在比对时可能产生补充比对(supplementary alignments)
上述片段会在 EPI2ME 工作流程中由 minimap2 自动过滤,但若您使用其他分析工具,则需将其纳入分析考量。
16S 引物设计
本试剂盒所含的 16S 引物未表现出易形成二聚体或发夹结构的倾向。







