Chemistry Technical Document (CHTD_500_v1_revAT_17Feb2026)
TechnicalDocument
V CHTD_500_v1_revAT_17Feb2026
FOR RESEARCH USE ONLY
Contents
Introduction to library preparation chemistry
Available kits
- 3. Ligation-based sequencing kits
- 4. Rapid-based sequencing kits
- 5. RNA and cDNA sequencing kits
- 6. Barcoding kits
- 7. Expansion packs
Sample input and recommendations
Making the most of your flow cell
Legacy kits
- 10. Ligation-based sequencing kits
- 11. Rapid-based sequencing kits
- 12. RNA and cDNA sequencing kits
- 13. Barcoding kits
Sequences and reagents
1. Introduction to library preparation chemistry
Library preparation is the conversion of a DNA or RNA sample to a suitable format for sequencing on Oxford Nanopore Technologies devices. The flow cells used for sequencing the samples contain ion-permeable nanopores embedded in an electrically-resistant membrane enabling an ionic current to pass through the nanopore when a voltage is applied across the membrane. This creates a measurable current that is disrupted when a strand of DNA or RNA passes through the nanopore. The disruption of current is measured and is used to identify the bases passing through the nanopore. Modified bases can also be identified as the nucleic acids are directly sequenced. This means PCR is not required, preventing PCR bias or polymerase error. However, PCR is available to use with our kits to generate more input or repair template damage.
DNA and RNA libraries are prepared by attaching sequencing adapters to strand ends, using either ligation-based or rapid chemistry methods. The sequencing adapters are oligonucleotides that are loaded with a motor protein. The motor protein associates with the nanopore in the flow cell and controls the DNA or RNA strand movement through the nanopores at a defined speed. A hydrophobic tether is also used to localise the template to the membrane into closer contact with the nanopores, improving sensitivity by approximately 10,000 fold.
There are two types of chemistry used in our sequencing kits:
- ligation-based chemistry: The sequencing adapter is ligated onto the DNA ends during library preparation.
- rapid-based chemistry: The sequencing adapter is attached onto the DNA without ligation enzymes, when the transposase cuts the DNA. In some kits, adaptation can also be performed by PCR.
Find out more information about our sequencing kits in this document.
2. Store terminology
Throughout the Community and Store, we use the following terminology to explain the capabilities of our kits:
Sample:
The starting material from which DNA or RNA is extracted from and used as input in a library preparation protocol.
DNA/RNA sample:
A collection of DNA or RNA fragments to be used as input in a library preparation protocol.
Library:
A collection of DNA or RNA fragments that have sequencing adapters attached in preparation for sequencing on a flow cell. This can be a collection of fragments from one sample or multiple DNA/RNA samples that have been barcoded and pooled together.
Barcodes:
A known sequence attached to a DNA fragment of a sample during library preparation, enabling users to combine multiple samples into a single library pool. Before the samples are pooled together, each sample must be uniquely and individually barcoded. The multiplexed library is sequenced and demultiplexed using MinKNOW for identification of the individual samples based on the known barcode sequences.
For example, six samples can be individually barcoded and pooled together using the Rapid Barcoding Kit 24 V14 (SQK-RBK114.24) before sequencing on a single flow cell. The samples are then demultiplexed by MinKNOW, allowing for analysis of individual samples based on their barcodes.
Reactions:
The number of times a kit can be used for either a library preparation, flow cell priming or as extra reagents to make full use of a sequencing kit.
For example, the Native Barcoding Kit 96 V14 (SQK-NBD114.96) can be used to sequence six libraries of 48 barcoded DNA samples. To make use of the remaining barcodes, a Native Barcoding Auxiliary Kit V14 (EXP-NBA114) is required to provide more library preparation reagents, along with a Sequencing Auxiliary Vials V14 (EXP-AUX003) expansion to provide the flow cell priming reagents.
Pack size:
The number of times a kit can be used to prepare a reaction. For example, the Flow Cell Priming Kit V14 (EXP-FLP004) has a pack size of six reactions, meaning enough reagents are supplied to prime six flow cells.
For the sequencing kits, pack size refers to the number of libraries and number of flow cells that can be primed for sequencing using the reagents supplied in the kit. For example, the Rapid Sequencing Kit V14 (SQK-RAD114) has a pack size of six reactions, that means the kit can prepare six libraries and contains enough reagents to prime six flow cells for sequencing.
For the barcoding kits, pack size also refers to the number of barcoded libraries and number of flow cells that can be primed for sequencing using the reagents supplied in the kit. However, for the kits containing 96 barcodes, up to 12 barcoded libraries can be prepared and sequenced, depending on how many DNA samples are barcoded. The below options outline how to maximise the kit for either barcodes or flow cell priming reagents:
- 3 libraries of 96 barcodes
- 6 libraries of 48 barcodes
- 12 libraries of 24 barcodes
To maximise the use of unused barcodes, the relevant expansion packs are available on the store.
Product phases
All our products fit into a product phase that is displayed on the store to make it clear where the product is in its lifecycle and degree of availability, warranty, and change notification. Product warranties and change notification periods increase through each phase. All products receive regular software and other platform updates that are rigourously checked and tested before release. These tests become more formalised as the products progress.
Product phases have no impact on ordering processes. They are simply there to provide users with guidance on how rapidly we expect to be applying changes to the product and accuracy of shipping dates in store.
3. Ligation-based sequencing kits
What is ligation-based sequencing?
Ligation-based chemistry is the method used to attach the sequencing adapter (by ligation) to the DNA ends during library preparation using ligation enzymes.
Our ligation sequencing kits are optimised for output and accuracy. These kits are the most popular kits and require double-stranded DNA as input, including gDNA, amplicons, or cDNA.
Our current available ligation sequencing kits are:
- Ligation Sequencing Kit V14 (SQK-LSK114)
- Ligation Sequencing Kit XL V14 (SQK-LSK114-XL)
- Native Barcoding Kit 24 V14 (SQK-NBD114.24)
- Native Barcoding Kit 96 V14 (SQK-NBD114.96)
- PCR Barcoding Expansion 1-12 (EXP-PBC001)
- PCR Barcoding Expansion 1-96 (EXP-PBC096)
- Multiplex Ligation Sequencing Kit V14 XL (SQK-MLK114.96-XL)
Below, we outline the sample input requirements and library preparation workflows for our kits. For information on the barcoding kits, please see the Barcoding kits section.
Sample input recommendations for the ligation-based sequencing kits
Before starting library preparation using the Ligation Sequencing Kit, it is important to make sure that you are using the correct amount of starting material for a successful sequencing experiment. After DNA extraction, we recommend quantifying your DNA samples.
For information on how to quantify the mass of DNA samples for library preparation input, please refer to the Sample input and recommendations section.
Depending on fragment lengths, we recommend different starting inputs for the Ligation Sequencing Kit V14 (SQK-LSK114) with R10.4.1 flow cells.
Note: These input recommendations are for our standard protocols and may vary depending on whether a protocol is written for a specific application.
| Samples | Input |
|---|---|
| Short fragment samples (<1 kb) | 200 fmol |
| Fragment samples (1–10 kb) | 100–200 fmol |
| Long fragment samples (>10 kb) | 1 µg |
Molar quantification is difficult and unreliable for samples consisting of long fragments and we have found that a starting input of 1 µg is almost always sufficient for optimal pore occupancy. The graph below illustrates pore occupancy when using different mass inputs of sample.
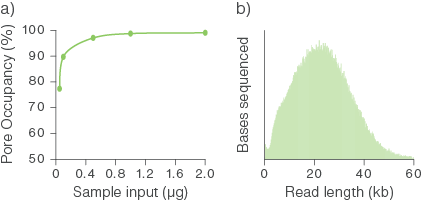
Figure 1. Relationship between sample input for library preparation and pore occupancy.
Sequencing libraries were prepared using various quantities of starting inputs of a genomic DNA sample comprising of long fragments and sequenced on PromethION Flow Cells. (a) A starting input of ~0.5 to 1 µg of gDNA was sufficient to achieve optimal pore occupancy. (b) The fragment length distribution of the sequenced library.
If you use less than 1 µg in your library preparation, you may have a reduced yield, meaning you will have fewer DNA strands with sequencing adapters (adapted DNA) on each end which may negatively affect sequencing output. This is because optimal pore occupancy may not be reached and it may deteriorate faster as the pores will not always be sequencing, compromising output. For more information on pore occupancy, please see the Sample input and recommendations section.
When starting with as little as 100 ng of high molecular weight (HMW) DNA, we have observed outputs of >50 Gb from R10.4.1 PromethION Flow Cells (Figure 2b). As you decrease input below 100 ng, pore occupancy significantly deteriorates, and we recommend considering amplification (by PCR or using Multiple Displacement Amplification) to generate more template. Shearing HMW DNA using Covaris g-TUBE or MegaRuptor® increases the strands of DNA for low input samples. For further information, see the Sample input and recommendations section.
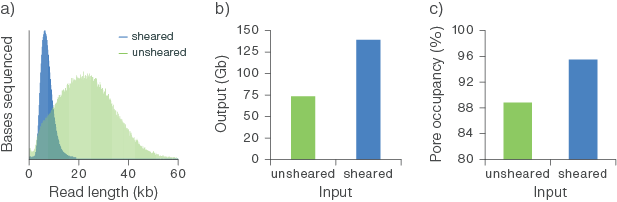 Figure 2. Sequencing output from a starting input of 100 ng of sheared and unsheared high molecular weight (HMW) human genomic DNA.
Figure 2. Sequencing output from a starting input of 100 ng of sheared and unsheared high molecular weight (HMW) human genomic DNA.
The gDNA was extracted from whole blood using the Puregene Blood Kit (Qiagen) and sheared using g-TUBE (Covaris). Libraries were prepared for sequencing with the Ligation Sequencing Kit V14 (SQK-LSK114) using 100 ng of sheared and unsheared template DNA. The libraries were sequenced on PromethION Flow Cells. (a) Shearing the input gDNA decreased the read lengths observed following sequencing. (b) The flow cell output (Gb) obtained from 100 ng of gDNA input was increased by shearing. (b-c) Shearing the gDNA increased the efficiency of pore occupancy, leading to a higher output from the flow cell.
Ligation Sequencing Kit V14
The Ligation Sequencing Kit V14 (SQK-LSK114) is our current ligation-based sequencing kit optimised to achieve sequencing accuracies of over 99% (Q20+), with high output on our latest nanopore: R10.4.1.
This Kit 14 upgrade includes previous updates such as higher capture rate of DNA to enable lower flow cell loading amounts, and fuel fix technology. Note, due to the higher capture of the adapter, it is important to follow the flow cell loading recommendations in the protocols.
The library preparation is simple and highly versatile, accommodating any double-stranded DNA sample input of 200 fmol for short fragment libraries (<1 kb), 100‒200 fmol for fragment libraries (1‒10 kb) or 1 µg of long fragment libraries (>10 kb). Fragment length can be controlled by optional fragmentation or size selection methods which can be found in our DNA/RNA handling page.
This kit is also compatible with upstream processes such as targeted enrichment by sequence capture and whole-genome amplification.
Details of our previous Ligation Sequencing kits are available in the Legacy kit section of this document.
Workflow:
The library preparation involves two enzymatic steps to prepare the DNA ends for sequencing. The first enzymatic step repairs any damage in the DNA molecules, such as nicks and generates uniform ends with 5’ phosphates and 3’ adenine overhangs. The second enzymatic step ligates the sequencing adapters that have complementary thymine tails with the dA-tailed template.
This kit has a standard protocol and multiple associated protocols. Due to its multiple applications, this kit can be used for:
• Ligation sequencing DNA V14 (SQK-LSK114)
• Ligation sequencing amplicons V14 (SQK-LSK114)
• Ligation sequencing gDNA - Lambda control V14 (SQK-LSK114)
• Ligation sequencing V14 - low input by PCR (SQK-LSK114 with EXP-PCA001)
• Ligation sequencing V14 - PCR barcoding (SQK-LSK114 with EXP-PBC001 or EXP-PBC096)
• Ligation sequencing V14 - Direct cDNA sequencing (SQK-LSK114)
• Human cfDNA singleplex sequencing from blood using SQK-LSK114
• Reduced representation methylation sequencing (RRMS) from cells using SQK-LSK114
• Human variation sequencing from 30kb extracted cell line samples using SQK-LSK114
• Human variation sequencing from 15kb extracted cell line samples using SQK-LSK114
• Human variation sequencing from 10kb extracted cell line samples using SQK-LSK114
• Human variation sequencing from 30kb extracted blood samples using SQK-LSK114
• Human variation sequencing from 15kb extracted blood samples using SQK-LSK114
• Human variation sequencing from saliva samples using SQK-LSK114
• Human variation sequencing from buccal swab samples using SQK-LSK114
• Ligation sequencing gDNA V14 - whole genome amplification (SQK-LSK114)
• Single-cell transcriptomics sequencing from 3’ cDNA prepared with 10x Genomics using SQK-LSK114 and EXP-PCA001
• Single-cell transcriptomics sequencing from 5’ cDNA prepared with 10x Genomics using SQK-LSK114
• Spatial transcriptomics sequencing from 3’ cDNA prepared with 10x Genomics using SQK-LSK114 and EXP-PCA001
• Chromatin accessibility sequencing from cell samples using SQK-LSK114
• 24-hour genome: end-to-end workflow from blood to analysis
Ligation Sequencing Kit XL
The Ligation Sequencing Kit V14 XL (SQK-LSK114-XL) is a scaled-up version of the Ligation Sequencing Kit V14 and contains larger quantities of the same components.
In this kit, there are sufficient reagents to generate 48 sequencing libraries and is recommended for users who would like to process multiple samples simultaneously, either with a multichannel pipette or a liquid handling robot.
4. Rapid-based sequencing kits
What is rapid-based sequencing?
Rapid-based chemistry is the method that attaches the sequencing adapter (rapid attachment) to the DNA ends. Typically, in the transposase step, the DNA is cut and the adapter is attached at the same time without any ligation enzymes. However, in the barcoding kits, the barcodes are attached during the transposase step and the sequencing adapters are attached in a later step using rapid attachment. It is also worth noting that in the PCR-based kits, adaptation is completed by PCR.
These kits are optimised for speed and simplicity or for applications where turnaround times are more critical or laboratory equipment is limited.
When compared with the Ligation Sequencing Kit V14, the sequencing output from the Rapid Sequencing Kit V14 is slightly reduced due to the lack of purification steps and the presence of transposases; both of which may cause pore blockages, preventing the pore from sequencing adapted DNA (Figure 3). Read length is also slightly reduced due to the requisite transposase fragmentation.
As the Rapid Sequencing Kit focuses on a rapid library preparation, certain steps, such as bead purification, are omitted and may reduce sequencing yield in some cases. For further information on improving flow cell performance, please refer to the Flow cell performance in the Sample input and recommendations section.
Figure 3. Comparison of throughputs from DNA libraries prepared using the Ligation Sequencing and Rapid Sequencing Kits.
The graph illustrates typical output achieved with the two kits.
Our current available rapid sequencing kits are:
- Rapid Sequencing Kit V14 (SQK-RAD114)
- Ultra-Long DNA Sequencing Kit V14 (SQK-ULK114)
- Rapid Barcoding Kit 24 V14 (SQK-RBK114.24)
- Rapid Barcoding Kit 96 V14 (SQK-RBK114.96)
- Rapid PCR Barcoding Kit (SQK-RPB114.24)
- 16S Barcoding Kit 24 V14 (SQK-16S114.24)
- Microbial Amplicon Barcoding Kit 24 V14 (SQK-MAB114.24)*
* Please note that the Microbial Amplicon Barcoding Kit 24 V14 (SQK-MAB114.24) utilises the rapid-based chemistry for adapter attachment. However, this is not transposase-based and utilises our new Amplicon Barcoding chemistry for multiplexing.
Below, we outline the sample input requirements and library preparation workflows for these kits. For information on the barcoding kits, please see the Barcoding kits section.
Sample input recommendations for the rapid-based sequencing kits
Before starting library preparation using the Rapid Sequencing Kit, it is important to make sure that you are using the correct amount of starting material for a successful sequencing experiment. After DNA extraction, we recommend quantifying your DNA samples.
For information on how to quantify the mass of DNA samples for library preparation input, please refer to the Sample input and recommendations section.
For the Rapid Sequencing Kit V14 (SQK-RAD114), a reduced starting input of 100 ng HMW gDNA containing long fragments is recommended. Using less than 100 ng gDNA or samples with shorter fragments can compromise sequencing output, as the yield from library preparation will be reduced. Where only lower inputs are available, we recommend using the Rapid PCR Barcoding Kit 24 V14 (SQK-RPB114.24) to increase the number of template molecules.
Please refer to the Sample input and recommendations section of this document for further information about sample input and how to improve library quality.
Data output from the flow cell is influenced by the amount and quality of the input DNA sample fragmented by the fixed amount of transposase in the sequencing kit to generate tagged fragments.
- To generate long fragments:
- Add more than the recommended starting input as there will be fewer cuts per molecule.
- Long fragments of DNA must be present initially in the sample input.
- To generate short fragments:
- Add less than the recommended starting input as there will be more cuts per molecule.
Due to the simple nature of the workflow and the fact that little sample manipulation is required (e.g. minimal pipetting steps and no clean-up steps), some very long reads can be achieved with this kit, despite the required transposase fragmentation. However, in order for long reads to be observed in sequencing, long fragments need to be present in the sample in the first place. To generate ultra-long reads, we recommend using the Ultra-Long DNA Sequencing Kit V14 (SQK-ULK114), which uses rapid-based chemistry. Further information on this sequencing kit is available below.
Rapid Sequencing Kit V14
The Rapid Sequencing Kit V14 (SQK-RAD114) is our newest rapid-based sequencing kit optimised for speed and simplicity, using limited laboratory equipment, and a lower starting input of 100 ng HMW gDNA. This kit has been upgraded to use our newest Kit 14 chemistry, which includes improved modal raw read sequencing accuracies with higher output on our latest nanopore: R10.4.1. This Kit 14 upgrade also includes updates such as the higher capture rate of DNA to enable lower flow cell loading amounts, and fuel fix technology. Note, due to the higher capture of the adapter, it is important to follow the flow cell loading recommendations in the protocols.
Due to the minimal sample manipulation required when using this kit, very long reads can be generated despite the requisite transposase fragmentation, as long as there are very long DNA fragments present in the input sample. However, for ultra-long reads, we recommend using the Ultra-Long DNA Sequencing Kit V14.
Workflow:
The library preparation uses transposase-based fragmentation of the gDNA input whilst simultaneously tagging the fragmented template with transposase adapter sequences. Post-transposition, sequencing adapters are then attached to the transposase adapters in an enzyme-free reaction.
Ultra-Long DNA Sequencing Kit V14
This is our updated Ultra-Long DNA Sequencing Kit using rapid-based Kit 14 chemistry to sequence ultra-long reads. The kit includes new reagents to simplify library preparation and to improve DNA precipitation and recovery. Due to the simple workflow, where minimal pipetting is required during library preparation, very long reads can be achieved.
This kit requires ultra high molecular weight (uHMW) gDNA to be extracted. In our protocol, we recommend using the NEB Monarch® HMW DNA Extraction Kit for Tissue (T3060) to extract the uHMW gDNA from either frozen cells or whole blood. After gDNA extraction, a diluted fragmentation mix containing transposases is added to the extracted gDNA to fragment and simultaneously tag the fragmented template with transposase adapter sequences. Post-transposition, sequencing adapters are then attached to the transposase adapters in an enzyme-free reaction. After an overnight elution with the Precipitation Star (PS), the library is ready for sequencing.
5. RNA and cDNA sequencing kits
Introduction to RNA and cDNA sequencing
Using nanopore technology, RNA can be directly sequenced without sequencing complementary DNA (cDNA) intermediates, enabling the exploration of the attributes of native RNA, such as base modifications. Our Direct RNA Sequencing Kit (SQK-RNA004) has improved sequencing outputs and accuracy on our newly released RNA flow cells.
We have also developed kits to sequence cDNA, which is a reverse-transcribed copy of native mRNA. Therefore, characteristics of cDNA can be exploited to select for fully reverse-transcribed cDNA molecules, using the cDNA-PCR Sequencing Kits.
Our current RNA and cDNA kits available are:
- Direct RNA Sequencing Kit (SQK-RNA004)
- Direct RNA Sequencing Kit XL (SQK-RNA004-XL)
- cDNA-PCR Sequencing Kit V14 (SQK-PCS114)
- cDNA-PCR Barcoding Kit V14 (SQK-PCB114.24)
Below, we outline the sample input requirements and library preparation workflows for these kits. For information on the barcoding kits, please see the Barcoding kits section.
Sample input recommendations for the RNA and cDNA sequencing kits
For our RNA and cDNA sequencing kits, poly(A)-tailed mRNA is required as input as the tails are used for adapter attachment and primer annealing. However, total RNA can be used as input to the library preparation as the primers and adapters used can select for the poly(A) tails. However, using total RNA requires a larger amount of input, as mRNA typically does not make up a large proportion of total RNA.
To overcome the issue with total RNA input, poly(A) polymerase can be used to polyadenylate the RNA sample, as described in the Polyadenylation of non-poly(A) transcripts using E. coli poly(A) polymerase document. Poly(A)-tailed mRNA can also be enriched using different methods, however, regardless of methods used, there may be a slight bias towards molecules with longer poly(A) tails during library preparation as longer tails may be better targets for selection protocols. Therefore, users should be mindful of which enrichment methods are most appropriate for their needs.
To ensure high-quality sample preparation, RNA samples should be free from contaminants. If total RNA is used, an RNA Integrity Number >7 is recommended before enriching the sample using poly(A) selection. A clean RNase-free environment is crucial with good RNase-cleaning practices to prevent RNA degradation. Further information is available in the RNA contaminants and RNA stability documents.
Direct RNA Sequencing Kits
The Direct RNA Sequencing Kit (SQK-RNA004) is our current RNA kit with improved outputs and accuracy compared to the previous version of this kit. Improvements to this kit include a faster motor protein, a new RNA-specific reader pore and an optimised library preparation kit.
We also have a Direct RNA Sequencing Kit XL (SQK-RNA004-XL) which is a scaled-up version of the Direct RNA Sequencing Kit containing larger quantities of the same components. In this kit, there are sufficient reagents to generate 48 sequencing libraries and is recommended for users who would like to process multiple samples simultaneously.
An input of 300 ng of poly(A) tailed RNA or 1 µg of total RNA is required. Other possible RNA inputs includes eukaryotic mRNA, viral RNA with a poly(A) tail, or any RNA prepared with a poly(A) tailing kit.
Workflow
Note: Only the RNA strand, not the reverse transcribed strand, is sequenced.
The library preparation prepares the RNA for sequencing by ligating a reverse transcription adapter to the RNA ends before performing reverse transcription to synthesise a complementary strand. This is to stabilise the RNA against secondary structure formation. However, only the RNA strand is sequenced. If reverse transcription is bypassed, the workflow is shortened to 30 minutes but sequencing output is reduced; this is likely due to an RNA tertiary structure blocking the pores. The sequencing adapters supplied in the kit are attached to the ends of the RNA-cDNA hybrid for sequencing.
cDNA-PCR Sequencing Kit V14
The cDNA-PCR Sequencing Kit (SQK-PCS114) is our updated cDNA sequencing kit and is recommended for users starting with a low input of 10 ng of poly(A) tailed RNA or 500 ng of total RNA, avoiding the need to poly(A) select the template molecules.
This kit can be used for the identification and quantification of full-length transcripts. Isoforms, splice variants, and fusion transcripts can also be sequenced for analysis.
The Kit 14 chemistry upgrade in this kit reduces the level of free sequencing adapter and improves sequencing accuracies and output. Other updates include the addition of a cDNA RT adapter and RT primer to reduce transcript overlaps during the reverse transcription step to enable the measurement of the poly(A) tail length and a unique molecular identifier (UMI) for the identification of splice variants.
Workflow:
The protocol starts with a reverse transcription step to prepare full-length cDNA from the input RNA and incorporates the UMI. During reverse transcription, the poly(dT) reverse transcription adapter is ligated to the 3’ terminal poly(A) tail of the template molecule. The bottom strand of the adapter is removed and a reverse transcription primer is annealed, anchoring the start of transcription to include the entire 3’ terminal poly(A) tail. Then a strand-switching primer, containing a UMI, is added during reverse transcription, allowing strand switching to occur and generate a full-length cDNA strand. This is tagged with universal sequences on both ends.
Note: The reverse transcriptase inhibits downstream PCR and the enzymes must be heat-inactivated and the reverse transcribed sample should be split across four PCR reactions to dilute the inhibitors. This is to allow the amplification of cDNA with maximum efficiency, without losing sensitivity in the next step.
Once the full-length cDNA is prepared, PCR amplification is performed and rapid attachment primers are added to the cDNA simultaneously. Finally, rapid sequencing adapters are attached to the primers for sequencing.
Using this kit, specific transcripts can be selected if one or both ends of the target cDNA are known:
- Both target ends are known: Reverse-transcribe the entire template molecule and use selective primers to anneal to both ends of the cDNA before carrying out PCR with the sequence-specific primers.
- Only one end of the target is known: Reverse-transcribe the template molecule and use selective primers to anneal to the known end of the cDNA with universal primers on the unknown end. Then carry out PCR with the sequence-specific primers for the known end of the target molecule and a universal primer for the unknown end.
A specific transcript can also be selected by altering the reverse transcription primer to replace the poly(dT) sequence with a sequence-specific primer so only the transcript of interest will be reverse-transcribed. This reverse-transcribed and strand-switched product is then amplified with the universal primers in the PCR-cDNA Sequencing Kit.
The cDNA-PCR Barcoding Kit (SQK-PCB114.24) is the barcoding version of this kit and can be found in the Barcoding kits section.
6. Barcoding kits
Introduction to barcoding kits
The barcoding kits are designed to allow the pooling and running of multiple libraries on our flow cell by attaching a known DNA sequence (barcode) to your samples within a pooled library. The pooled library is then sequenced in a single run and MinKNOW uses the barcodes to demultiplex the library during a sequencing run. The barcodes have been carefully designed and extensively purified to minimise cross-talk.
We recommend using our barcoding kits:
- To reduce cost per sample.
- For more efficient use of flow cells when less data per sample is required than the total amount of data that can be generated from a single flow cell.
- For optimal re-use of a flow cell when used with the Flow Cell Wash Kit, as any barcoded residual reads on a flow cell will be identified between barcoded samples.
Note: As more samples are multiplexed, less data per sample is generated.
Multiplexing samples onto one flow cell can reduce the cost per sample for a user. In the Table below, we illustrate how barcoding 1 to 96 samples onto a single flow cell can alter the cost per sample.
Note: These costs are an example and do not reflect our current store prices.
| Barcodes | 0 | 6 | 12 | 24 | 48 | 96 |
|---|---|---|---|---|---|---|
| Flow cell price | $500 | $500 | $500 | $500 | $500 | $500 |
| Library price | $99 | $99 | $99 | $99 | $99 | $99 |
| Barcode price | - | $18.75 | $37.50 | $75 | $150 | $300 |
| Price per sample | $599 | $102.95 | $53.04 | $28.08 | $15.60 | $9.36 |
There are four types of barcoding kits:
- Ligation-based barcoding kits
- Ligation-based PCR barcoding kits
- Rapid chemistry-based barcoding kits
- Rapid chemistry-based PCR barcoding kits
Our current barcoding kits have been updated to combine all the reagents required for library preparation, barcoding and flow cell priming into a kit.
All our kits with 96 barcodes have been developed to allow flexibility in the number of samples that can be multiplexed. Users can prepare multiplexed libraries using various options without extra expansion packs:
- 12 libraries containing 24 barcodes
- 6 libraries containing 48 barcodes
- 3 libraries containing 96 barcodes
Expansion packs for library preparation and flow cell priming reagents are available to make use of any remaining barcodes in a kit.
The barcode sequences are available at the end of this document in the section Sequences and reagents.
Ligation-based barcoding kits
Native Barcoding Kit 24 and 96 V14
There are two standalone native barcoding kits and one expansion:
- Native Barcoding Kit 24 V14 (SQK-NBD114.24)
- Native Barcoding Kit 96 V14 (SQK-NBD114.96)
- Native Barcoding Auxiliary Kit V14 (EXP-NBA114)
The Native Barcoding Kits 24 and 96 V14 contain all the required reagents for barcoding 24 or 96 samples, preparing libraries for sequencing and flow cell priming. The kits are optimised to achieve sequencing accuracies of over 99% (Q20+), with high output.
The Native Barcoding Kits are recommended for users who want to multiplex their samples with a PCR-free method to preserve base modifications. There are up to 24 or 96 unique barcodes which can be used for gDNA or amplicons. The barcoding kits are optimised to generate maximum output without the need for PCR.
These kits require double-stranded DNA as input with the following recommendations below. Due to the higher capture of the adapter, it is important to follow the flow cell loading recommendations in the protocols.
| Kit | Input requirements |
|---|---|
| Native Barcoding Kit 24 V14 (SQK-NBD114.24) for barcoding up to 24 samples | • ≤4 barcodes, 1000 ng of gDNA per sample • >4 barcodes, 400 ng of gDNA per sample • 200 fmol (130 ng for 1 kb amplicons) DNA per sample |
| Native Barcoding Kit 96 V14 (SQK-NBD114.96) for barcoding up to 96 samples | • 400 ng of gDNA per sample • 200 fmol (130 ng for 1 kb amplicons) DNA per sample |
The Native Barcoding Auxiliary Kit V14 (EXP-NBA114) contains additional library preparation reagents for use with the Native Barcoding Kits for another 12 reactions. For further information on our available expansions, please see the Expansion packs section.
Workflow
The DNA is repaired and the ends dA-tailed in preparation for the dT-tailed native barcode ligation. After barcoding, the samples are pooled together and sequencing adapters are ligated to the barcode ends for sequencing.
Multiplex Ligation Sequencing Kit XL V14
This kit (SQK-MLK114.96-XL) provides 96 barcodes for the high output and PCR-free, low-plex sequencing of double-stranded DNA. This kit is recommended for low-plex whole genome sequencing. All required reagents for barcoding up to 96 samples, preparing libraries for sequencing and flow cell loading are included, using our Kit 14 chemistry.
We recommend using 1 µg gDNA per sample to sequence two samples per flow cell. This results in the sequencing of up to 96 samples across 48 flow cells. Three samples across two flow cells can also be used to sequence up to 96 samples across 64 flow cells to maximise sequencing output.
This kit uses our native barcodes and is compatible with the Native Barcoding Auxiliary Kit V14 (EXP-NBA114) for additional library preparation reagents.
Workflow
The library preparation method is similar to the Ligation Sequencing Kit protocol; DNA ends are repaired and dA-tailed in preparation for the dT-tailed native barcode ligation. After barcoding, the samples are pooled together and the sequencing adapters are ligated to the barcode ends for sequencing.
Ligation-based PCR barcoding kits
PCR Barcoding Expansion 1-12 and 1-96
For low starting inputs, there are two PCR barcoding expansion packs with 12 or 96 barcodes; PCR Barcoding Expansion 1-12 (EXP-PBC001) and PCR Barcoding Expansion 1-96 (EXP-PBC096). These barcoding expansions are available to use in combination with our Ligation Sequencing Kit, to enable the pooling and running of multiple PCR amplified sequencing libraries for low inputs of gDNA and amplicon. They include adapters and up to 96 forward and reverse PCR primers with the same barcode in both forward and reverse primers.
An input of 100 ng of gDNA is required per sample. For amplicons,100 ng of first-round PCR product with tailed primers is required per sample.
Note: Amplicons have to be prepared in advance for barcoding, as the first round of amplification is not included in the library preparation protocol.
Details of recommendations are outlined in the Ligation sequencing V14 - PCR barcoding (SQK-LSK114 with EXP-PBC001 or EXP-PBC096) protocol.
Workflow
Depending on your input, preparing the sample for the library preparation step will vary.
For low gDNA inputs, you will need to prepare your DNA ends for PCR adapter ligation and then attach the barcoding adapters to the DNA ends.
For low amplicon inputs, you will need to perform a round of PCR to incorporate tailed primers. The amplicon samples receiving the same barcode can be pooled before the barcoding PCR step is performed.
The barcoding PCR step is the same across all sample inputs following which the pooled barcoded samples are end-prepped for the sequencing adapters to ligate to the barcodes before sequencing.
Dual barcoding protocol
To perform massive, parallel sequencing of up to 2,304 samples of gDNA or amplicons on a single flow cell, the PCR Barcoding Expansion 1-96 (EXP-PBC096) can be combined with the Native Barcoding Kit 24 V14 (SQK-NBD114.24). The dual barcoding protocol combines both kits to dual barcode the libraries for pooling and sequencing on a single flow cell.
We recommend dual barcoding for users wishing to sequence more than 96 samples or want to sort libraries into multiple categories as each barcode can be used to denote a different sample feature. The inner barcode is a PCR barcode and the outer barcode is a native barcode.
An input of 100 ng of gDNA is required per sample. For amplicons,100 ng of first-round PCR product with tailed primers is required per sample.
Note: Amplicons have to be prepared in advance for barcoding, as the first round of amplification is not included in the library preparation protocol.
Details of recommendations are outlined in the Ligation sequencing DNA V14 - dual barcoding (SQK-NBD114.24 with EXP-PBC096) protocol.
Workflow
The ends of the DNA input are first prepared for PCR adapter ligation after which PCR is performed with barcoded primers to barcode up to 96 samples. The 96 samples are pooled into a single library and this is repeated to create 24 pools of 96 barcoded samples. For each pool, the DNA ends are prepared for native barcode ligation. After barcoding, the dual barcoded libraries are combined into a single pool. Sequencing adapters are ligated to the native barcodes in the pool, before sequencing on a single flow cell.
Rapid chemistry-based barcoding kits
Rapid Barcoding Kit 24 and 96
There are two standalone rapid barcoding kits and one expansion using Kit 14 chemistry:
- Rapid Barcoding Kit 24 V14 (SQK-RBK114.24)
- Rapid Barcoding Kit 96 V14 (SQK-RBK114.96)
- Rapid Adapter Auxiliary V14 (EXP-RAA114)
The Rapid Barcoding Kit 24 V14 (SQK-RBK114.24) and Rapid Barcoding Kit 96 V14 (SQK-RBK114.96) are both standalone kits for barcoding and sequencing up to 24 or 96 samples, respectively, using a library preparation optimised for speed and simplicity, and requiring minimal laboratory equipment. For users requiring high accuracy and output, we recommend the Native Barcoding Kit 24 or 96 V14 (SQK-NBD114.24 and SQK-NBD114.96).
A sample input of 200 ng of gDNA is required for each sample to be barcoded and sequenced.
Workflow
The DNA input undergoes a transposase step to fragment the sample while attaching rapid barcodes to the ends simultaneously. The samples can be pooled and a bead clean-up performed to remove any excess transposases in the reaction. The sequencing adapters are then attached to the barcodes and the library can be sequenced.
Rapid chemistry-based PCR barcoding kits
Rapid PCR Barcoding Kit 24 V14
The Rapid PCR Barcoding Kit 24 V14 (SQK-RPB114.24) is the fastest and simplest method for preparing barcoded libraries starting with low inputs of gDNA. PCR amplification is performed to increase the number of templates for sequencing. This kit has been upgraded to use Kit 14 chemistry and the number of barcodes available in the kit has been increased to 24. The Kit 14 upgrade includes improved raw read sequencing accuracies and higher output. The barcode primers in this kit are shipped at 10 µM in vial format or 1 µM in plate format.
A low starting input of 1‒5 ng of gDNA per sample is required.
Note: The DNA must be at least 4 kb in length to ensure correct tagmentation and PCR amplification.
Workflow
The gDNA is prepared for sequencing by undergoing tagmentation to fragment the DNA. PCR is performed to amplify the DNA and attach the rapid barcode primers to the DNA ends. After barcoding, the samples are pooled and the rapid sequencing adapter is attached to the ends of the DNA samples for sequencing.
16S Barcoding Kit 24 V14
This is our updated 16S kit (SQK-16S114.24) using our latest Kit 14 chemistry to amplify the entire ~1,500 bp 16S rRNA gene from extracted gDNA. With our chemistry upgrade, sequencing accuracies and output are improved and a lower flow cell loading amount is required. The number of barcodes have also been increased to enable barcoding of up to 24 samples into a single library. The barcodes in our 16S rapid-based kit are shipped at 1 µM.
We recommend this kit for rapid sequencing of the 16S rRNA gene for bacterial identification in samples. By narrowing down to a specific region of interest, all organisms in a sample can be screened, without sequencing unnecessary regions of the genome. This will improve the efficiency of identification by reducing time and costs.
An input of 10 ng of gDNA per sample is required.
Workflow
The gDNA undergoes PCR amplification with 16S barcode primers (27F and 1492R) to amplify the 16S gene present in the samples. Barcoded samples are pooled before a bead clean-up. Then rapid adapters are attached to the DNA ends in the pooled sample for sequencing.
Amplicon barcoding chemistry kits
Microbial Amplicon Barcoding Kit
There is one Amplicon Barcoding chemistry-based kit:
- Microbial Amplicon Barcoding Kit 24 V14 (SQK-MAB114.24)
The Microbial Amplicon Barcoding Kit 24 V14 (SQK-MAB114.24) is a standalone kit for barcoding and sequencing up to 24 samples, using a library preparation that is fragmentation-free, while optimising for speed and simplicity, and requires minimal laboratory equipment. The amplicon barcodes contain 5’ tags which facilitate the ligase-free attachment of Rapid Sequencing Adapters.
This kit contains updated primers that enable full-length sequencing of 16S rRNA for bacterial profiling and ITS for fungal profiling from diverse sample types.
A sample input of 10 ng of gDNA is required for each sample to be barcoded and sequenced.
Workflow
The gDNA samples are amplified using the 16S or ITS primers supplied in the kit. An equimolar concentration of each amplicon sample is taken forward following PCR, and the amplicon barcodes are added to each sample, providing fragmentation-free barcode attachment. The amplicon barcodes are inactivated using Proteinase K, and the barcoded samples are pooled before performing a sample clean-up. The rapid sequencing adapters are then attached to the amplicon barcodes before sequencing.
Primer design, PCR and sequencing considerations:
A full list of the primer sequences for both the 16S primers (16S) and ITS primers (ITS) can be found in this downloadable document:
Our primer schemes are aligned with industry standards and provide coverage for the entire 16S rRNA and ITS rRNA gene respectively. However, the primers used bind within regions that are not 100% conserved across species and therefore we are unable to guarantee amplification for all species.
Please ensure you run a control experiment to determine if your targeted species will be amplified using the primers provided in this kit. If you require further support, contact us via support@nanoporetech.com.
cDNA barcoding kit
cDNA-PCR Barcoding Kit V14
This is our recently updated cDNA barcoding kit (SQK-PCB114.24) to barcode up to 24 RNA samples for sequencing of the cDNA. Identification and qualtification of full-length transcripts can be performed with this kit and isoforms, splice variants, and fusion transcripts can be sequenced. The barcode primers in this kit are shipped at 10 µM in vial format or 1 µM in plate format.
The Kit 14 chemistry upgrade in this kit reduces the level of free sequencing adapter and improves sequencing accuracies and output. Other updates include the addition of a cDNA RT adapter and RT primer to reduce transcript overlaps during the reverse transcription step to enable the measurement of the poly(A) tail length and a unique molecular identifier (UMI) for the identification of splice variants.
A low input of 10 ng of enriched RNA per sample is required which can be poly(A) tailed or ribodepleted RNA. Total RNA can also be used but 500 ng is required per sample.
Workflow
The RNA undergoes a reverse transcription step to prepare full-length cDNA from the input RNA and incorporates the UMI. During reverse transcription, the poly(dT) reverse transcription adapter is ligated to the 3’ terminal poly(A) tail of the template molecule. The bottom strand of the adapter is removed and a reverse transcription primer is annealed, anchoring the start of transcription to include the entire 3’ terminal poly(A) tail. Then a strand-switching primer, containing a UMI, is added during reverse transcription, allowing strand switching to occur and generate a full-length cDNA strand. This is tagged with universal sequences on both ends.
Note: The reverse transcriptase inhibits downstream PCR and the enzymes must be heat-inactivated and the reverse transcribed sample must be split across four PCR reactions to dilute the inhibitors. This is to allow the amplification of cDNA with maximum efficiency, without losing sensitivity in the next step.
Once the full-length cDNA is prepared, PCR amplification is performed and rapid barcode primers are attached to the cDNA samples. After barcoding, the samples are pooled and rapid sequencing adapters are attached for sequencing.
7. Expansion packs
Introduction to expansion packs
Expansion packs are available to be used alongside the sequencing kits and include extra reagents or barcodes for more efficient use of our sequencing kits.
Below is an overview of our current expansion packs, however, the store expansion pack page also includes additional simple expansions containing additional reagents.
Sequencing Auxiliary Vials and XL
This Sequencing Auxiliary Vials V14 (EXP-AUX003) is recommended for users who want to split a library across multiple flow cells, need to add more library to an existing run, or wish to top up a flow cell with an additional library. This expansion contains sufficient reagents for six reactions when used with our V14 kits (except for the Ultra-Long DNA Sequencing Kit V14), and includes additional flow cell priming reagents. The Sequencing Auxiliary Vials V14 XL (EXP-AUX003-XL) is a scaled up version of the expansion, providing enough reagents for 64 reactions.
Note: The Ultra-Long DNA Sequencing Kit V14 (SQK-ULK114) is not compatible with the Sequencing Auxiliary Vials as it requires specific flow cell priming reagents which are not included in this expansion.
Flow Cell Wash Kit and XL
This expansion (EXP-WSH004) is recommended for users who want to run multiple sequencing libraries on the same flow cell and is compatible with R10.4.1 flow cells. It can also be used when a flow cell has accumulated a high number of ‘unavailable’ pores during a run and users want to revert these pores to the available state.
This expansion provides a highly effective means of removing a library that has been loaded onto a flow cell. Once the flow cell is washed, it is available immediately for use again or can be stored. There are sufficient reagents for six flow cell washes. The Flow Cell Wash Kit XL (EXP-WSH004-XL) is a scaled up version of the kit, providing enough reagents for 48 flow cell washes.
We recommend barcoding samples when running multiple samples sequentially to allow for filtering of sequences remaining from previous runs as there may be some residual DNA on the flow cell.
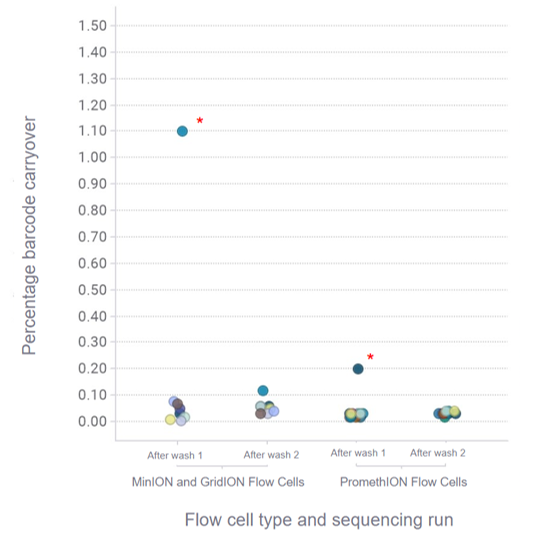
Figure 4. Barcode carryover between sequencing runs.
Three libraries containing 4 barcodes each were sequenced sequentially following a flow cell wash with EXP-WSH004 between the sequencing runs. Libraries were prepared with Lambda DNA (LMD from the Control Expansion Kit (EXP-CTL001)) using the Rapid Barcoding Kit 96 V14 (SQK-RBK114.96) and tested across 8 MinION and GridION R10.4.1 Flow Cells, and 15 PromethION R10.4.1 Flow Cells. Each flow cell load comprised of an equal mixture of 4 barcodes per sequencing run. Sequencing runs were performed for 45 minutes. For MinION and GridION Flow Cells, 14 out of 16 runs showed <0.1% barcode carryover between sequencing runs following a flow cell wash, with an outlier in run 2 being due to user error. For PromethION Flow Cells, 29 out of 30 runs showed <0.1% barcode carryover between sequencing runs following a flow cell wash.
Flow Cell Priming Kit and XL
We have released a separate expansion (EXP-FLP004) containing just flow cell priming reagents. These are to be used alongside our sequencing kits to prime a MinION/GridION and PromethION Flow Cells, or to add more library to a flow cell during a sequencing experiment for another six reactions. The Flow Cell Priming Kit XL (EXP-FLP004-XL) is a scaled up version of the kit, providing enough reagents for 48 reactions.
Note: The Ultra-Long DNA Sequencing Kit V14 (SQK-ULK114) is not compatible with the Flow Cell Priming Kits as it requires specific flow cell priming reagents which are not included in this kit.
Short Fragment Eliminator Kit
The Short Fragment Eliminator Kit (EXP-SFE001) is designed for size selection of high molecular weight DNA samples by depleting short fragments of <25 kb. It is recommended for users who want to enrich their libraries for long fragments without using any external third-party reagents. This kit is compatible with all Oxford Nanopore DNA sequencing kits.
Please refer to the following protocol: Removal of short fragments with the Short Fragment Eliminator Kit.
Ultra-Long Auxiliary Vials
The Ultra-Long Auxiliary Vials (EXP-ULA001) is recommended for users who want to carry out additional flow cell loads from existing ultra-long libraries prepared using the protocol for the Ultra-Long DNA Sequencing Kit V14 (SQK-ULK114). This expansion contains enough reagents for an additional 12 reactions.
PCR Expansion
This expansion pack (EXP-PCA001) contains PCR Primers (PRM) and PCR Adapters (PCA) and is recommended for users needing reagents for a PCR-based sequencing experiment with the Ligation Sequencing Kit V14 (SQK-LSK114). This kit is recommended for low starting inputs that require PCR for preparing enough DNA for sequencing.
For more information, please see the relevant protocol:
Native Barcoding Auxiliary Kit V14
This expansion (EXP-NBA114) contains additional library preparation reagents for use with unused barcodes in the Native Barcoding Kits 24 and 96 V14 (SQK-NBD114.24 and SQK-NBD114.96). There are sufficient reagents for another 12 reactions supplied with this kit.
Note: This kit must be used in conjunction with the Sequencing Auxiliary Vials V14 expansion (EXP-AUX003) to provide the flow cell priming reagents.
Note: This product contains AMPure XP reagent manufactured by Beckman Coulter, Inc. and can be stored at -20°C with the kit, without detriment to reagent stability.
Rapid Adapter Auxiliary V14
This expansion (EXP-RAA114) provides additional Rapid Adapter (RA) and Adapter Buffer (ADB) to maximise any unused barcodes in the Rapid Barcoding Kit 24 and 96 V14 (SQK-RBK114.24 and SQK-RBK114.96).
Note: This kit must be used in conjunction with the Sequencing Auxiliary Vials V14 expansion (EXP-AUX003) to provide the flow cell priming reagents.
PCR Barcoding Expansion
The PCR Barcoding Expansion 1-12 (EXP-PBC001) and 1-96 (EXP-PBC096) are designed to allow the pooling and running together of multiple sequencing libraries. The expansions include adapters and forward and reverse PCR primers from Oxford Nanopore, with the same barcode in both forward and reverse primers.
For EXP-PBC001, there are 12 unique barcodes and enough of the required adapters to make 6 libraries per barcode. For EXP-PBC096 (plate format), there are 96 unique barcodes and enough of the required adapters to make 10 libraries per barcode. The barcodes have been carefully designed and extensively purified to minimise the opportunity for cross-talk.
There are currently two protocols which use these expansions:
- Ligation sequencing V14 - PCR Barcoding (SQK-LSK114 with EXP-PBC001 or EXP-PBC096)
- Ligation sequencing DNA V14 - dual barcoding (SQK-NBD114.24 with EXP-PBC096)
Note: If you are barcoding cDNA or amplicons, these have to be pre-made in advance.
The protocols listed can be adapted and optimised for specific experimental requirements.
Midnight RT PCR Expansion
The Midnight RT PCR Expansion (EXP-MRT001) has been developed to work in conjunction with the Rapid Barcoding Kit 96 V14 (SQK-RBK114.96) to enable PCR tiling of overlapping 1.2 kb amplicons across the SARS-CoV-2 genome.
The primers provided in this expansion were initially developed by Freed et al., 2020, using the Primal Scheme. These primers have been updated as new mutations were identified in the SARS-CoV-2 lineages and we will work closely with the community (including the entire ARTIC Network, Nikki Freed, Olin Silander, Josh Quick, John Tyson, Nick Loman and many more) to rapidly respond and ensure that Oxford Nanopore protocols are always kept up-to-date, providing the best performance for robust SARS-CoV-2 whole genome sequencing.
For this expansion, an input of extracted RNA in 10 mM of Tris-HCl, pH 8.0 is required.
Workflow
The extracted RNA is reverse transcribed with random hexamers and the samples are split into pools A and B for tiled PCR. The primer pools are combined with their associated sample, making sure to not combine different samples together. The DNA samples are then fragmented using a transposase and the rapid barcodes are attached to the fragment ends simultaneously. After barcoding, the samples can be combined into a single pool and a bead clean-up performed to remove any excess barcodes and transposases before sequencing on a flow cell.
8. Sample input and recommendations
The importance of good quality library
To make the most of a flow cell, load the correct amount of good quality library recommended in the appropriate protocol for your sequencing kit. A good quality library is made up of sample molecules that have sequencing adapters ligated at both ends. To achieve high sequencing output, the flow cell membrane must not contain air bubbles which can damage the pores, and pore blocking should be at a minimum. There must also be enough library input to ensure the pores are always sequencing with minimal idle time between sequencing strands. In this section, we discuss how to achieve this.
How much to load on a flow cell is dependent on library preparation protocols, starting input and fragment length. For standard library preparation using the Ligation Sequencing Kit, we recommend a starting input of ~100–200 fmol for fragments of 1–10 kb, or 1 µg for longer fragments of >10 kb. Starting input will also differ depending on user needs and starting input quality. For example, to generate ultra-long reads, ultra-long fragments must be present in the starting input sample.
It is important to load the recommended amount of library because this will affect data output. For example, not adding enough library onto the flow cell will impact pore occupancy as the optimal number of pores will not be sequencing DNA/RNA strands, reducing sequencing output.
Please refer to the Making the most of your flow cell for further information on how to interpret and use real-time data to improve sequencing output.
Quantifying DNA sample mass input for library preparation
Before starting library preparation, it is important to make sure that you are using the correct amount of starting material for a successful sequencing experiment. After DNA extraction, we recommend quantifying your DNA samples using the methods outlined in the Table below:
| Quantification | DNA fragment size | Method |
|---|---|---|
| Mass | n/a | Qubit™ Fluorometer using Qubit™ dsDNA Broad Range (BR) Quantification Assay (Thermo Fisher Scientific) |
| Size | <10 kb >10 kb | 2100 Bioanalyzer (Agilent) or equivalent, or pulsed-field gel electrophoresis Femto Pulse System (Agilent) or pulsed-field gel electrophoresis |
| Purity | n/a | NanoDrop™ 2000 Spectrophotometer (Thermo Fisher Scientific) or equivalent |
To quantify the mass of DNA samples, we recommend the Qubit dsDNA BR Assay Kit. Both mass and length measurements are needed for molar quantification of short-fragment samples.
To assess length, we recommend gel-based analysis or the Agilent 2100 Bioanalyzer (or equivalent). While molar quantification is not required for samples that comprise of long fragments, we still recommend measuring the length of your sample. This is to ensure your sample contains long fragments if you are interested in generating long reads. For this, we recommend using the pulsed-field gel electrophoresis or the Agilent Femto Pulse (the Agilent 2100 Bioanalyzer is not suitable for measuring the length of molecules >10 kb).
The kit used for DNA extraction can affect the DNA fragment length and subsequently the sequencing read length as shown in Figure 5.
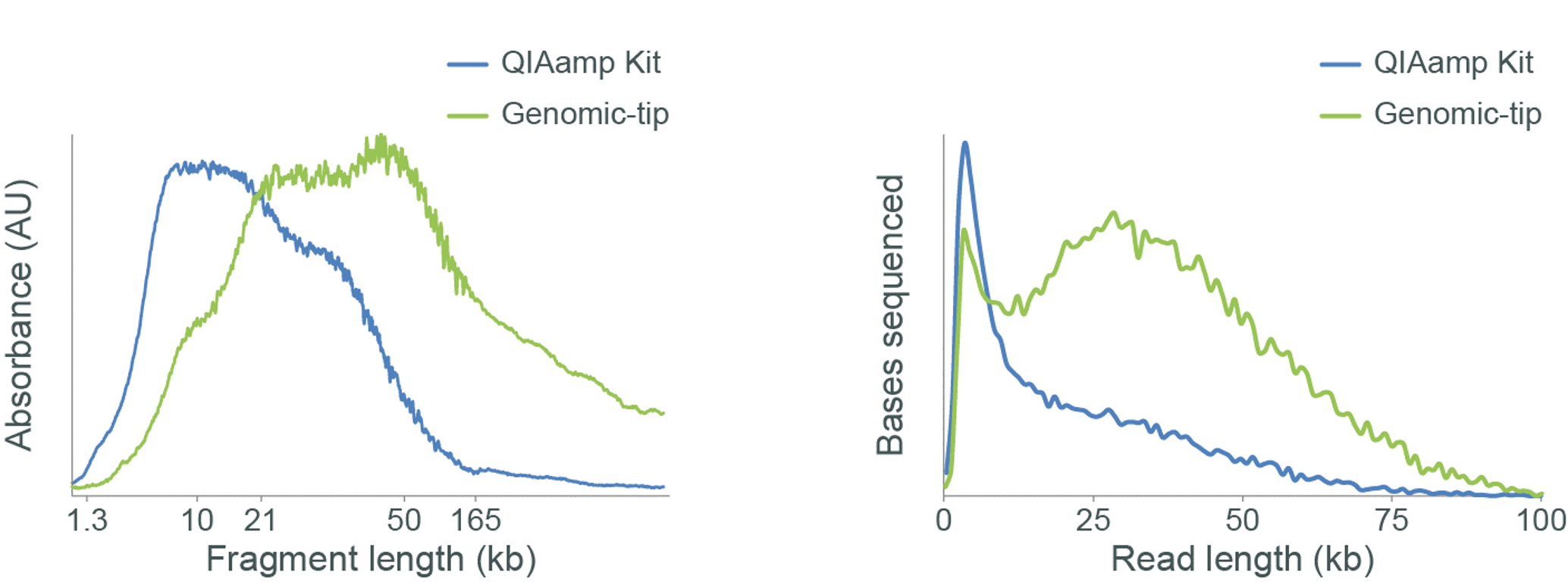
Figure 5. Assessing the fragment length of DNA extracted from samples using different kits.
Genomic DNA (gDNA) was extracted from rabbit blood using either the QIAamp DNA Blood Midi Kit or Genomic-tips from Qiagen. The extracted gDNA was analysed using the Agilent Femto Pulse System (left). Libraries were prepared using the Ligation Sequencing Kit V14 (SQK-LSK114) and sequenced on a MinION. The read length distributions for the sequenced libraries are shown (right). The Femto Pulse System shows that the QIAamp extracted sample is comprised mainly of short fragments (<10 kb), and this is reflected in the sequencing data. However, the Genomic-tip extracted sample is shown to contain a higher proportion of longer fragments, therefore producing a higher proportion of longer reads on the MinION.
Purity of DNA samples
It is also recommended to measure the purity of extracted DNA. DNA extraction methods often involve chemicals that can inhibit enzymatic activity, including those enzymes in library preparation.
To measure the presence of certain contaminants, the Nanodrop 2000 Spectrophotometer can be used. Pure DNA gives an A260/A280 ratio of ~1.8. A ratio lower than this can be indicative of a contaminant such as phenol. A A260/A230 ratio lower than 2.0–2.2 can also be indicative of phenol contamination but can also indicate carryover of guanidine or carbohydrate. However, while Nanodrop readings are a good place to start, it is not the only indicator of purity and subsequent DNA performance. Details of the effects of certain contaminants on the efficiency of the Ligation Sequencing Kit can be found in the DNA contaminants and RNA contaminants documents.
Pore occupancy and library quality
A good quality library is also required and refers to the extent of how well the DNA has been adapted with sequencing adapters. Molecules without sequencing adapters cannot be sequenced and diminish library quality. Libraries that consist of molecules with adapters and tethers on both ends are more potent than those with only one end modified.
The less library you load, the fewer “threadable ends” will be present to be captured by the pores. Therefore, the pores will be “searching” for molecules for longer, and if the pores are not always sequencing, then output could be compromised. It is important to note that we do not observe a linear relationship between input onto the flow cell and sequencing output, but loading less could give reduced output. Conversely, loading more library does not guarantee good performance.
Determinants of read length
We have found that long reads can be achieved with both ligation and rapid based library preparation methods, as read length reflects input fragment length and is not platform-limited. For example, fragmenting input DNA with g-TUBE results in a narrower distribution of read lengths (Figure 6). In some protocols, we recommend fragmenting input DNA to ease handling and quantification or to ensure multiple samples are uniform in size. We have also validated several methods of enrichment for long reads, including using SPRI beads and the Short Fragment Eliminator Kit (EXP-SFE001) that are located in the Size Selection page.
 Figure 6. Read length from DNA prepared with or without fragmentation or size selection.
Figure 6. Read length from DNA prepared with or without fragmentation or size selection.
DNA was either sheared using g-TUBE or size selected using BluePippin. Libraries were prepared using the Rapid Sequencing Kit (SQK-RAD114). (Top left) Read length using the sequencing kit protocol with the Fragmentation Mix. (Top right) Read length with g-TUBE shearing only. (Bottom left) Read length with no fragmentation or size selection. (Bottom right) Read length using BluePippin for size selection.
Size selection
Size selection is an optional step to enrich for long fragments or deplete short fragments in the library input. In order to obtain long reads, long fragments must be present in the extracted DNA sample. All extraction techniques that we recommend yield some short fragments (<10 kb) and even if they do not appear to be prevalent during quantification, it is likely that you will observe some short reads.
SPRI size selection is used in many Oxford Nanopore Technologies library preparation protocols as this technique is effective at enriching for fragments above ~1.5 kb and can slightly shift read length towards the longer end. The Short Fragment Eliminator Kit (EXP-SFE001) may be used to remove fragments of <25 kb and almost completely removing fragments under 10 kb. The size selection buffer (SSB) promotes semi-selective precipitation and enrichment of molecules above ~10 kb and the BluePippin has a tuneable cut-off limit of up to 40 kb. These methods effectively remove shorter fragments and boost read N50 without compromising data output. To view our protocols for size selection, please refer to the Size selection page.
To demonstrate their performance, gDNA was size selected using the following methods: SPRI bead selection, SFE Kit, and BluePippin (set to enrich for molecules >40 kb) (Figure 7). The data below shows the read N50 observed and the output generated for each of the libraries. Very little difference in output was observed over the course of the 24 hour sequencing run for SPRI and SFE Kit size-selected libraries, compared with the control. However, an accumulation of pores in the unavailable state were observed throughout the course of the run for the BluePippin size selected library, so a flow cell wash was performed after ~16 hours and more library loaded before recommencing the run.
Figure 7. Effect of various size selection methods on the read N50 and sequencing output of DNA.
Three µg of gDNA was size selected using either SPRI bead selection or the Short Fragment Eliminator Kit (EXP-SFE001) and 5 µg gDNA was size selected using BluePippin. The Ligation Sequencing Kit V14 (SQK-LSK114) was used to prepare 1 µg of each size selected DNA for sequencing, and the libraries were run on a MinION. A library where no size selection had been previously performed was also sequenced as a control. The read N50 values for the control, SPRI bead, SFE Kit, and BluePippin size selected libraries were ~20 kb, ~25 kb, ~35 kb, and ~41 kb, respectively.
Fragment length
The pores in a flow cell can sequence all sizes of fragments and read lengths are only limited by template molecule starting length. Nanopores capture and process both long and short DNA and RNA strands. However, the length of fragments can affect output as the concentration of free DNA ends will be different. For example, short fragments have a higher concentration of DNA ends that will increase pore occupancy and output. This is because there are more threadable ends for the pores to capture.
Libraries composed of long fragments typically have lower data outputs than short fragments as there are fewer molecules available for sequencing and they take longer to be captured by the pore. This also results in more pores not sequencing due to the lack of pore occupancy. If size selection is performed as part of the library preparation, this can lower output further due to fragments outside a particular range being excluded.
Fragmentation in a Covaris g-TUBE has been found to yield the highest output with no size selection. Therefore, we recommend using this fragmentation method in many protocols to optimise for highest output yield.

Figure 8. Sequencing output from DNA prepared with or without fragmentation or size selection.
DNA was either sheared using g-TUBE or size selected using BluePippin. Libraries were prepared using the Rapid Sequencing Kit (SQK-RAD114). (Blue bar) DNA was sheared using the Fragmentation Mix from the sequencing kit. The g-TUBE fragmentated DNA was shown to yield the highest output.
Flow cell performance
Flow cell output is governed by various factors, including DNA/RNA library input, loading amounts, and pore blocking. Below, we review how controlling the input material size distribution by fragmentation impacts these factors and what our recommendations are to generate the best data, based on experimental aims.
Increasing read N50
It has been observed that some shearing of gDNA samples can lead to an increase in observed read length. This seems counterintuitive — how can breaking up the DNA fragments give rise to longer reads? It has been suggested that certain fragments may be so long that they become “lost” during the library preparation and therefore are not observed, leaving only the short fragments (for example, the very longest fragments may not efficiently bind to, or elute from the AMPure beads used after end-prep or ligation). Light shearing, for example using the Megaruptor, can break up the very longest molecules into chunks that the library preparation can more readily process, leading to increased read N50s.
This approach is suggested where samples appear to have very high molecular weight in gel or Femto Pulse analysis but the observed read length N50 is <15 kb. Users within the Nanopore Community have also attempted other shearing methods to increase read lengths. However, the more aggressive the fragmentation, the higher the risk of over-fragmenting, leading to a reduction in observed read lengths.
 Figure 9. Effect of shearing on fragment length and read N50 of high molecular weight genomic DNA.
Figure 9. Effect of shearing on fragment length and read N50 of high molecular weight genomic DNA.
Human gDNA was extracted from cells in culture with the aim of recovering the longest possible fragments. (A) The resulting gDNA was sheared with Megaruptor® 3 (Diagenode) using a selection of shearing speeds and analysed using the Femto Pulse System. (B) Sequencing libraries were prepared using the Ligation Sequencing Kit V14 (SQK-LSK114) and run on the MinION. Read N50 values were recorded using the sequencing data. The read length distribution of the unsheared input shows that most of the DNA was above 100 kb, with a spike at 165 kb (area where fragments become compressed). However, this does not correspond to a high read N50 value following sequencing. The lowest shearing speed had minimal or no effect on the fragment length distribution or the read N50, suggesting unsuccessful fragmentation. However, increasing the shearing speed to 23–30 did show successful fragmentation and led to an increase in read N50. Increasing the shearing speed further, led to over-fragmentation and a drop in the read N50.
Input amount and pore occupancy
Loading too much or too little library can compromise flow cell performance. If sufficient starting material is unavailable, users can start with lower inputs, however we have found that data output can drop at lower inputs as there are insufficient molecules available to maximise pore occupancy. Fragmenting the sample (for example using a Covaris g-TUBE or Megaruptor®) can be used to increase the number of molecules/ends to thread into the nanopores. This increases pore occupancy and recovers the output.
It is worth noting that fragmenting DNA to boost the output can mean it may not be possible to achieve ultra-long reads.

Figure 10. Relationship between sample input and sequencing output for sheared and unsheared genomic DNA.
(A) As the input of unsheared gDNA in the library preparation decreased below 500 ng, the percentage pore occupancy decreased. Shearing the lower input samples using g-TUBE increased the pore occupancy with the sheared libraries on the flow cell when compared with the unsheared libraries. (B) Shearing the samples with g-TUBE had an impact on the read length distribution.
If you have <100 ng of DNA, we advise performing PCR to increase the amount of DNA available for sequencing. There are library preparation kits and protocols available specifically for PCR-based preparation:
Ligation sequencing V14 - low input by PCR (SQK-LSK114 with EXP-PCA001)
Rapid sequencing DNA - PCR Barcoding Kit 24 V14 (SQK-RPB114.24)
The choice of kit is dependent on the priorities of the user.
If inputs below the protocol recommendations are used, the sequencing output may be negatively affected due to the low pore occupancy.
To maximise sequencing output, it is important that the pores are kept filled with DNA to minimise the time that they are “idle” in-between strands. This metric can be monitored by viewing the pore occupancy graph on the MinKNOW UI. For more information, see the MinKNOW protocol.
Pore blocking
Pore blocking is another factor that can affect flow cell output. During a sequencing run, pores can become “blocked”, preventing the pore from accepting a new strand for sequencing or continuing to sequence the occupying strand. Such blocks are detected by the MinKNOW software, which changes the channel state from “single pore” to “unavailable”. For the duration of a blockage, the pore does not acquire any sequencing data. MinKNOW attempts to drive out whatever has blocked the pore by reversing the voltage. The unblocking scheme is progressive, increasing the duration of the voltage reversal until the blockage is cleared. Most of the time (~98%), attempts to unblock a pore are successful and it reverts to the single pore state, where it is available to accept new strands and continue sequencing. However, in a minority of cases, the progressive unblocking scheme will not be able to recover a blocked pore. If a pore becomes terminally blocked and cannot be recovered, a new pore is swapped in from a different well in the channel, if available. Typically, a blockade occurs every 250–500 kb and is successfully removed ~98% of the time (in other words, around 1 in 50 attempts will be unsuccessful). This gives an average output of ~10–20 Mb per pore, which for a flow cell containing ~1500 pores could lead to a total output of ~15–30 Gb (note, other factors may limit the actual output obtained). If there is an increase in the rate of blocking, then pores spend less time sequencing and more unblocks are triggered. If there are more unblocks, or if the success of unblocking decreases, then the rate at which pores are lost increases and total flow cell output is reduced.
To determine if fragment length plays a role in the rate of pore blocking, DNA extracted from human cells grown in culture was sheared with Megaruptor 2 (Figure 11) . We were not able to establish a relationship between read length and blocking rate, although we observed a decrease in the success rate of the unblock for the longer libraries, indicating that our unblocking scheme is less capable of removing blocks from longer fragments. Given this observation, if you obtain a low output, then some shearing of the sample could be performed to see if unblocking can be improved to help boost output.
 Figure 11. Effect of fragment length on blocking and unblocking of pores, and flow cell output.
Figure 11. Effect of fragment length on blocking and unblocking of pores, and flow cell output.
Extracted gDNA from a human cell line GM12878 was sheared with Megaruptor 2 using different shearing settings, or with g-TUBE. Libraries were then prepared using the Ligation Sequencing Kit V14 (SQK-LSK114) and sequenced on the MinION. (A) The read N50 values for the unsheared and sheared libraries. Note that some Megaruptor shearing produced a slightly elevated read N50 value. (B) The block frequency (kb/block) for the sheared libraries suggests that there is no correlation between the frequency at which blockades occurred and the length of the fragments that were being sequenced, at least for this sample. (C) As the fragment length of the library increased, a decrease in the rate of success of the unblock (number of blocks before a terminal block was encountered) was observed. (D) As a result of the decrease in unblock success with increased read lengths, it follows that the output from any pore (and hence whole flow cell) may also decrease with increased read length.
Recommendations for libraries with high levels of blocking:
- Fragment the starting DNA to increase pore occupancy.
- Flow cell washing may be used to unblock pores and revert the ‘unavailable’ state to ‘single pore’ state. The nuclease in the Flow Cell Wash Kit is able to digest all of the sample remaining on the flow cell and contaminants blocking the pores to increase output for more library to be loaded and sequenced.
9. Making the most of your flow cell
Overview of flow cells
The flow cell is the platform onto which a DNA/RNA library is loaded and sequenced. Sequencing devices can only use compatible flow cells. All our flow cells contain the proprietary sensor array, Application-Specific Integrated Circuit (ASIC) and nanopores for sequencing DNA/RNA libraries. The flow cell contains sufficient buffer to run for up to or more than 72 hours under optimal conditions depending on the flow cell. The user can choose to run continuously or run, stop, wash and load a new library until the buffer and nanopores are exhausted. This enables a single flow cell to be used multiple times before the nanopores are exhausted. An individual flow cell may also be used for different experiments.
The Table below shows the storage and minimum pore counts for the MinION/GridION and PromethION Flow Cells.
| Flow cell | MinION/GridION | PromethION |
|---|---|---|
| Device | MinION and GridION | PromethION |
| Storage and stability (unopened) | +2°C to +8°C: 12 weeks | +2°C to +8°C: 12 weeks |
| Minimum Pore Count | 800 | 5000 |
Nanopore technology allows for real-time assessment of sequencing experiments with the visualisation of run statistics on the MinKNOW UI. This makes it easier for the user to adjust sequencing parameters during a run setup and to make the most of a flow cell.
For libraries of high purity and quality, MinKNOW will be able to generate high data output. We recommend monitoring the pore activity plot to ensure pore occupancy remains high, as well as the pore scan plot to check whether there is a build up of unavailable channels. The translocation speed plot should also be checked to ensure sequencing speed remains between the median target. This is discussed in more depth further in the document.
We recommend live basecalling to allow the user to assess in real-time how the experiment is being conducted on the flow cell. However, the raw data can be saved and basecalled at a later date if the computer is struggling with real-time basecalling.
For further information on how to monitor a sequencing experiment, please refer to the MinKNOW protocol.
Flow cell wash
The Flow Cell Wash Kit (EXP-WSH004) contains a nuclease, DNase I, which is used to digest any remaining library on a flow cell for immediate reuse. The wash step is recommended when there is an accumulation of pores in the ‘Unavailable’ state.
In Figure 12 below, we have demonstrated that pores can be reverted to the 'Pore available' state by pausing sequencing and washing the flow cell with DNase I in the Flow Cell Wash Kit (EXP-WSH004). The asterisks indicate where sequencing has been paused and the flow cell washed.
Note: If the sequencing run is paused in MinKNOW for the flow cell wash, you will only see the restoration of sequencing pores after a new pore scan has been performed.
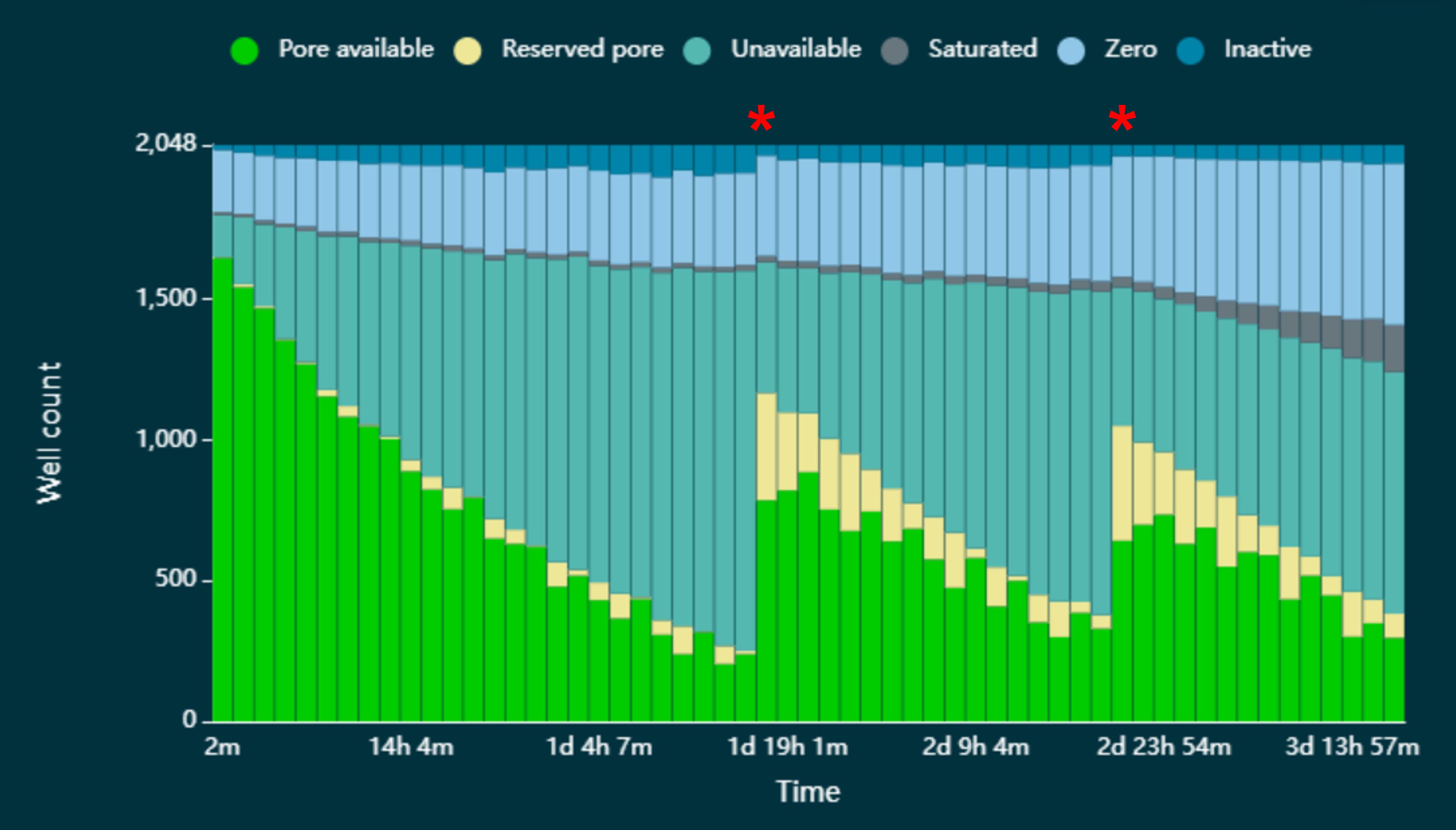
Figure 12. Pore states observed on a MinION/GridION Flow Cell before and after wash steps were performed.
The flow cell was loaded with a sequencing library that resulted in an accumulation of pores in the 'Unavailable' state, leading to a decrease in the rate of data acquisition. However, when each time a wash step was performed (indicated by red asterisk), a significant number of the pores that were lost to the 'Unavailable' state reverted to the 'Pore available' state and were available for sequencing once again.
The wash step is only recommended where sequencing channels are lost to the 'Unavailable' state. Where pores are in the ‘Saturated’ state, the wash step will not be able to revert them to the ‘Pore available’ state.
The flow cell wash kit allows multiple uses of a flow cell with different samples and the recovery of unavailable channels during an experiment. Samples can be multiplexed to reduce costs and many samples can be sequenced simultaneously on a single flow cell by using either a Native Barcoding Kit or a PCR Barcoding Expansion.
Note: The wash kit should remove 99% of the library. However, some residual DNA may remain on the flow cell. Therefore, we recommend barcoding the libraries when used in conjunction with the Flow Cell Wash Kit, to ensure reads from different libraries can be separated from each other.
RNA is also efficiently flushed out of the flow cell but it is not digested. The kit will wash off most of the library from the array and remove all adapter from the remaining sample, preventing it from being recaptured and sequenced. This will allow a subsequent library to be loaded.
The Flow Cell Wash Kit (EXP-WSH004) presents two options:
Wash and store the flow cell
- Allows for flow cells to be used for multiple experiments but not to be reused immediately after one experiment is complete.
Wash and reuse the flow cell straight away
- We recommend using the pause option in the MinKNOW UI to pause the experiment and to restart as soon as the wash is complete and the library is loaded.
- This is recommended when there is a build-up of ‘Unavailable’ pores.
- Note: When the Pause option is used, all data produced pre- and post-wash is stored in the same data file. Therefore, if changing the sample, be sure to use the Stop function rather than pausing, to ensure data is stored in a new file when sequencing is restarted.
To either pause or stop a sequencing run to wash a flow cell, navigate to the Experiments page and select either Pause or Stop in the run controls. This will open a dialogue box where you can select which run(s) to pause/stop.

Pore occupancy
Pore occupancy is driven by the number of sequenceable ends (sample molecules with sequencing adapters) loaded onto the flow cell. We recommend monitoring saturated and blocked pores which may reduce the output from a run if not dealt with appropriately.
During the sequencing experiment, the user can check various flow cell performance parameters that are shown in the MinKNOW UI. The channel states panel illustrates the real-time representation of each of the pores in a flow cell. The pore activity plot is a summary of time spent in each of the channel states during the sequencing run.
For detailed information, please refer to the MinKNOW protocol.
Channel states
The channel states panel gives an overview of the states the flow cell pores are in, to give the user an idea of how well the sequencing run is performing in real time. A good library will be indicated by a higher proportion of light green channels in "Sequencing" than are in "Pore available". The combination of "Sequencing" and "Pore available" indicates the number of active pores at any point in time. A low proportion of "Sequencing" channels will reduce the output of the run.

Figure 13. Example of a channel states panel for a sequencing run.
Clicking on the Show detailed tab reveals a more detailed array of channel states.
Pore activity plots
The pore activity plot summarises the channel states over time. Each bar shows the sum of all channel activity in a particular amount of time.
The graph populates over time, and can be used as a way to assess the quality of your sequencing experiment, and make an early decision whether to continue with the experiment or to stop the run.

Figure 14. Example of a pore activity plot.
Clicking on the Show detailed tab reveals a more detailed view of channel states.
Pore scan
The pore scan analyses pore status and selects the best performing pores for sequencing. We recommend triggering a pore scan after pausing or stopping an experiment, whether it is for reloading or washing a flow cell. This is to ensure all the nanopore channels are assessed most recently with the best selection of pores for sequencing.
The pore scan results can be used to assess sequencing efficiency with the duty time plot. If there is a high level of ‘Unavailable’ pores, we recommend washing the flow cell as there is blocking occurring which will reduce sequencing and data output. The rate of blocking is mainly due to the purity of the sample; low quality DNA/RNA or contamination from either extraction, sample prep or secondary structure can be some of the causes for an increase of pores in the 'Unavailable' state.
To start a pore scan, navigate to the Experiments page, and click Start pore scan in the run controls. A dialogue box will open where you can select the run(s) to be pore scanned. This will start a pore scan on the flow cell(s) in the selected run(s).

Read length histogram
This visualises the read lengths of the sequencing libraries in real-time and changes throughout the experiment. This can be used to assess if the sequencing library contains an expected sample if the read length distribution roughly matches the input library fragment sizes. If read length does not reflect the input library fragment sizes, the experiment can be stopped and the flow cell washed and stored whilst the library preparation can be optimised, preventing the waste of a flow cell.
Note: This graph will take slightly longer to populate for longer reads.
 Figure 15. Example of a read length histogram from a sequencing run.
Figure 15. Example of a read length histogram from a sequencing run.
For further information on how to monitor a sequencing experiment, please refer to the MinKNOW protocol.
Disk space plot
This allows the user to monitor the amount of data to be written onto the device memory during sequencing. If the disk space is running low during a run, the user can pause the experiment whilst making more disk space available and preserving the flow cell.
10. Ligation-based sequencing kits
Ligation-based sequencing kits
These are the previously available Ligation Sequencing Kits using our Kit 9, 10 and 12 chemistry:
- Ligation Sequencing Kit (SQK-LSK112)
- Ligation Sequencing Kit XL (SQK-LSK112-XL)
- Ligation Sequencing Kit (SQK-LSK110)
- Ligation Sequencing Kit (SQK-LSK109)
- Ligation Sequencing Kit XL (SQK-LSK109-XL)
- Cas9 Sequencing Kit (SQK-CS9109)
- Native Barcoding Kit 1-12 (EXP-NBD104)
- Native Barcoding Kit 13-24 (EXP-NBD114)
- Native Barcoding Kit 96 (EXP-NBD196)
- Multiplex Ligation Sequencing Kit XL (SQK-MLK111.96-XL)
Ligation Sequencing Kits
SQK-LSK112
The Ligation Sequencing Kit (SQK-LSK112) was our previous upgrade from the Ligation Sequencing Kit (SQK-LSK110) and is optimised for high accuracy. The upgrade included a new sequencing adapter (Adapter Mix H, AMX H) which improved accuracies to over 99% (Q20+), required lower flow cell loading amounts and contained the fuel fix technology to run long experiments without the need for fuel addition during a run.
Note: This kit (SQK-LSK112) is a legacy product and will soon be discontinued. We recommend all customers to upgrade to the latest chemistry for their relevant kit which is available on the Store. If customers require further support for any ongoing critical experiments using a Legacy product, please contact Technical Support via email: support@nanoporetech.com.
SQK-LSK110
The Ligation Sequencing Kit (SQK-LSK110) is an upgrade from the Ligation Sequencing Kit (SQK-LSK109) and is optimised for high output. The upgrade contains new fuel fix adapter (Adapter Mix F, or AMX-F) to use significantly less fuel during a sequencing run to improve sequencing output. We have also upgraded the loading beads to Loading Beads II (LBII) and Loading Solution (LS) for users who do not use loading beads for their experiments.
SQK-LSK109
This is a ligation-based sequencing kit to offer a flexible method of preparing sequencing libraries from dsDNA (e.g. gDNA, cDNA or amplicons) and is optimised to generate maximum output. Since release, this kit has undergone multiple upgrades and we recommend customers to upgrade to our latest chemistry to achieve the best results.
These are the previously available expansion packs available to work alongside the Ligation Sequencing Kits to enable users to barcode their samples for more efficient use of the kits.
Barcoding expansion packs available for the Ligation Sequencing Kits:
- Compatible with Ligation Sequencing Kit (SQK-LSK109):
- Native Barcoding Expansion 1-12 (EXP-NBD104)
- Native Barcoding Expansion 13-24 (EXP-NBD114)
- Native Barcoding Expansion 96 (EXP-NBD196)
Cas9 Sequencing Kit (SQK-CS0109)
The Cas9 Sequencing Kit (SQK-CS9109) uses ligation-based chemistry and is a fast and flexible way to target specific regions of interest within a genome without amplification, increasing the depth of coverage. This is particularly useful for areas not amenable to PCR, “dark” areas of the genome, or for users wishing to target specific regions but maintain DNA modifications. The Targeted, amplification-free DNA sequencing using CRISPR/Cas document explains how to design and order crRNA probes and shares best practices for performing targeted sequencing.
Using the recommended 5 µg high molecular weight gDNA, the DNA is dephosphorylated before adding Cas9 ribonucleoprotein particles (RNPs) with bound crRNA and tracrRNA to the gDNA for binding and cleavage of the Region of Interest (ROI). The cleavage by Cas9 reveals blunt ends with 5’ phosphates which are dA-tailed to prepare the blunt ends for barcode ligation. Sequencing adapters are then ligated to the Cas9-cut sides. Finally, a clean-up step is performed to remove excess adapters and the library is sequenced.
11. Rapid-based sequencing kits
Rapid-based kits
Oxford Nanopore has previously offered the following rapid-based sequencing kits:
- Rapid Sequencing Kit (SQK-RAD004)
- Rapid Barcoding Kit (SQK-RBK004)
- Ultra-Long DNA Sequencing Kit (SQK-ULK001)
- Field Sequencing Kit (SQK-LRK001)
- PCR Sequencing Kit (SQK-PSK004)
- PCR Barcoding Kit (SQK-PBK004):
- Rapid PCR Barcoding Kit (SQK-RPB004)
- 16S Barcoding Kit (SQK-RAB204)
- 16S Barcoding Kit 1-24 (SQK-16S024)
Rapid Sequencing Kit
SQK-RAD004
The Rapid Sequencing Kit (SQK-RAD004) is optimised for speed and simplicity, using limited laboratory equipment. The transposase-based workflow takes approximately 10 minutes to generate sequencing libraries from extracted gDNA in two steps. We recommend an input of 400 ng of HMW gDNA (>30 kb).
Ultra-Long DNA Sequencing Kit
As our transposase-based kits are relatively simple, very long reads can be achieved due to the minimal pipetting required during library preparation. The Ultra-Long DNA Sequencing Kit (SQK-ULK001) was developed using our rapid-based kit.
This kit requires ultra-high molecular weight (uHMW) gDNA to be extracted. In our protocol, we recommend using the NEB Monarch® HMW DNA Extraction Kit for Tissue (T3060) to extract the uHMW gDNA from either frozen cells or whole blood. After gDNA extraction, a diluted fragmentation mix containing transposases are added to the extracted gDNA to fragment and simultaneously tag the fragmented template with transposase adapter sequences. Post-transposition, sequencing adapters are then attached to the transposase adapters in an enzyme-free reaction. After an overnight elution with Elution Buffer, the library is ready for sequencing.
Field Sequencing Kit
The Field Sequencing Kit (SQK-LRK001) is recommended for those working in the field with limited access to cold storage and laboratory equipment. It is recommended to use ~400 ng high molecular weight gDNA as input for the library preparation. This kit uses a simple two-step protocol, using a transposase to simultaneously cleave the template molecules and attach tags to the cleaved ends. The Rapid Sequencing Adapters are then added to the tagged ends in approximately 10 minutes. This kit is stable at ambient temperatures for extended periods of time and many of the reagents are lyophilised.
PCR Sequencing Kit
The PCR Sequencing Kit (SQK-PSK004) is designed to prepare sequencing libraries when the available gDNA input is limited (<100 ng). We recommend starting with ~100 ng gDNA but less can be used. The gDNA is fragmented in a Covaris g-TUBE and the sheared ends are repaired and dA-tailed before adapters containing primer binding sites are ligated onto the prepared ends. An amplification step uses primers containing 5’ tags for the ligase-free attachment of Rapid Sequencing Adapters. Prior to PCR, this workflow takes approximately 50 minutes and post-PCR, the attachment of sequencing adapters takes about 10 minutes.
The barcoding version is the PCR Barcoding Kit (SQK-PBK004), enabling the multiplexing of up to 12 samples.
12. RNA and cDNA sequencing kits
RNA and cDNA sequencing kits
These are the previously available RNA and cDNA kits.
- cDNA-PCR Sequencing Kit (SQK-PCS109)
- cDNA-PCR Barcoding Kit (SQK-PCS111)
- Direct cDNA Sequencing Kit (SQK-DCS109)
- PCR-cDNA Barcoding Kit (SQK-PCB109)
- PCR-cDNA Barcoding Kit (SQK-PCB111.24)
- Direct RNA Sequencing Kit (SQK-RNA002)
Direct RNA Sequencing Kit
The Direct RNA Sequencing Kit (SQK-RNA002) is used to prepare any RNA with a 3’ poly(A) tail for sequencing. Other possible RNA input includes eukaryotic mRNA, viral RNA with a poly(A) tail, or any RNA prepared with a poly(A)-tailing kit. For RNA without a poly(A) tail, users can follow simple instructions to design their own custom adapter to ligate a specific terminal 3’ sequence like the 3’ of tRNA or 16S rRNA. We recommend an input of ~500 ng of poly(A) RNA template of >200 nucleotides in length, with no upper limit.
Note: Only the RNA strand, not the RT strand, is sequenced.
Only the native RNA passes through the nanopore, meaning the read length reflects the length of the RNA molecules in the sample. The first step of the workflow requires the ligation of a reverse transcription adapter which ligates to the 3’ poly(A) tail on the RNA template molecule. Next, the optional reverse transcription step can be carried out to generate cDNA for stabilisation against RNA secondary structure formation. However, only the RNA strand is sequenced. If reverse transcription is bypassed, the workflow is shortened to 30 minutes but there is a reduction in sequencing output which is likely due to an RNA tertiary structure blocking the pores. The sequencing adapters supplied in the kit are attached to the ends of the RNA-cDNA hybrid and the library is ready for sequencing.
cDNA-PCR Sequencing Kit
The cDNA-PCR Sequencing Kit (SQK-PCS111) has been to Kit 11 chemistry, which requires starting inputs of 4 ng poly(A) RNA or 200 ng of total RNA. We have included a new cDNA RT adapter (CRTA), Annealing Buffer (AB) and RT primer (RTP) to prime cDNA synthesis from the end of a transcript to reduce overlaps during the reverse transcription step and to allow users to measure poly(A) tail lengths. The Kit 11 upgrade includes a new Rapid Adapter T (RAP T), with two key features which includes higher capture rate and fuel fix technology for longer experiments without the need for fuel addition during the run.
Note: Reverse transcriptase inhibits downstream PCR. Therefore, the enzymes must be heat-inactivated and the reverse transcribed sample to be split across four PCR reactions to dilute the inhibitors. This is to allow the amplification of cDNA with maximum efficiency, without losing sensitivity.
The poly(dT) reverse transcription adapter is ligated to the 3’ terminal poly(A) tail of the template molecule. The bottom strand of the adapter is removed and a reverse transcription primer is annealed, anchoring the start of transcription to include the entire 3’ terminal poly(A) tail. Then a strand-switching primer containing a unique sequence (UMI) is added during reverse transcription, allowing strand switching to occur and generates a full-length cDNA strand with universal sequences on both ends. Next, PCR amplification is performed using primers with 5' tags and rapid attachment groups on the ends for adding the Rapid Sequencing Adapter (RAP T).
This kit can be used to select specific transcripts if one or both ends of the target are known:
- Both target ends are known: Reverse-transcribe the entire template molecule and use selective primers to anneal to both ends of the cDNA before carrying out PCR with the sequence-specific primers.
- Only one end of the target end is known: Reverse-transcribe the template molecule and use selective primers to anneal to the known end of the cDNA with universal primers on the unknown end. Then perform PCR with the sequence-specific primers for the known end of the target molecule and a universal primer for the unknown end.
A specific transcript can be selected by altering the reverse transcription primer to replace the poly(dT) sequence with a sequence-specific primer so only the transcript of interest will be reverse-transcribed. This reverse-transcribed and strand-switched product is then amplified with the universal primers in the PCR-cDNA Sequencing Kit.
PCR-cDNA Barcoding Kit (SQK-PCB111.24) is the barcoding version of this kit which can be found in the Barcoding kits section.
Direct cDNA Sequencing Kit
This kit is used to prepare cDNA for nanopore sequencing without using PCR. Preparing a library using the Direct cDNA Sequencing Kit (SQK-DCS109) takes approximately two hours longer than the cDNA-PCR Sequencing Kit (SQK-PCS109), and requires more third-party reagents. This is a PCR-free protocol and requires a higher input of 100 ng of poly(A)-tailed RNA but prevents PCR bias or polymerase error. However, data output is lower compared to the PCR version (PCR-cDNA Sequencing Kit). We recommend starting with 100 ng poly(A)-tailed RNA, or 70-200 ng of already prepared cDNA.
The first step of the workflow starts with annealing the reverse transcription primer and adding the strand-switching primer before performing reverse transcription to allow strand-switching to occur. An RNase cocktail is used to remove the RNA template strands before performing a second strand synthesis. This generates double-stranded cDNA which is treated similarly to gDNA. The fragment ends are dA-tailed before the dT-tailed sequencing adapter is ligated onto the fragments for sequencing on a flow cell.
13. Barcoding kits
Ligation-based barcoding kits
Native Barcoding Expansions 1-12, 13-24 and 96
For the Kit 9 Ligation Sequencing Kit (SQK-LSK109), there are three native barcoding expansions to enable barcoding:
- Native Barcoding Expansion 1-12 (EXP-NBD104)
- Native Barcoding Expansion 13-24 (EXP-NBD114)
- Native Barcoding Expansion 96 (EXP-NBD196)
These native barcoding expansions are recommended for users who want to multiplex their samples with a PCR-free method to preserve base modifications. There are up to 96 unique barcodes which can be used with the Ligation Sequencing Kits (SQK-LSK109) to sequence gDNA or cDNA with the Direct cDNA Sequencing Kit (SQK-DCS109). For more information about sequencing cDNA, please see the RNA and cDNA sequencing and kits section.
The barcode expansion packs are optimised to generate maximum output without the need for PCR. For highest data yields, we recommend the following inputs:
Native Barcoding Expansion 1-12 (EXP-NBD104) and 13-24 (EXP-NBD114) for barcoding up to 24 samples:
- 1 µg or 1.5-3 µg pure input DNA for R9.4.1 or R10.3 flow cells.
Native Barcoding Expansion 96 (EXP-NBD196) for barcoding up to 96 samples:
- ~400 ng DNA per barcode for R9.4.1 or R10.3 flow cells.
Multiplex Ligation Sequencing Kit XL
The Multiplex Ligation Sequencing Kit XL (SQK-MLK111.96-XL) is a standalone kit providing 96 unique barcodes to enable PCR-free multiplexing of dsDNA samples such as gDNA and amplicons. This kit provides an easier workflow to enable Whole Genome Sequencing (WGS) by enabling low-plex sequencing of 2-3 samples, allowing users to identify regions more common and others more prone to mutations.
The library preparation is similar to the Ligation Sequencing Kit protocol; DNA ends are repaired and dA-tailed using the NEBNext End Repair/dA-tailing module, and then a unique dT-tailed barcode adapter is ligated on the dA-tailed template. Barcoded samples are then pooled together. Each barcode adapter also has a cohesive end and this is used as a hook to ligate to the supplied sequencing adapters.
We recommend starting with 1 μg gDNA per sample and our manual protocol recommends sequencing two samples per flow cell enabling the sequencing of 96 samples across 48 flow cells. There is also an automated protocol on the Hamilton NGS STAR 96 (NGS STAR with Multi-Probe Head 96) to sequence either two samples per flow cell or three samples across two flow cells which is fully supported by Oxford Nanopore Technologies.
Rapid-based barcoding kits
Rapid Barcoding Kit
The Rapid Barcoding Kit (SQK-RBK004) can be used to barcode up to 12 samples quickly with limited laboratory equipment. It is recommended to use ~400 ng high molecular weight gDNA to generate barcoded libraries in 15 minutes using the simple two-step protocol.
This kit uses the transposase simultaneously cleaves template molecules and attaches barcoded tags to the cleaved ends. Barcoded samples are pooled and Rapid Sequencing Adapters are then added to the tagged ends.
Rapid-based PCR Barcoding Kits
PCR Barcoding Kit (SQK-PBK004):
The PCR Barcoding Kit (SQK-PBK004) is the multiplexing version of the PCR Sequencing Kit (SQK-PSK004), allowing up to 12 samples to be multiplexed on one flow cell when starting with low input. When the quantity of input DNA is low or users want to maximise output, we recommend using PCR to increase the amount of template molecules. This kit implements PCR and it is recommended to start with ~100 ng gDNA. However, less input can be used but it may be necessary to increase the number of PCR cycles.
The gDNA samples are fragmented in a Covaris g-TUBE and the sheared ends are repaired and dA-tailed before the adapters containing primer binding sites are ligated onto the prepared ends. This kit contains 12 primer pairs which are used to amplify each sample during PCR. After amplification, the Rapid Sequencing Adapters are attached to the pooled barcoded samples. Each primer pair contains a barcode and 5’ tags which facilitates the ligase-free attachment of the Rapid Sequencing Adapter. Prior to PCR, this workflow takes approximately 50 minutes and post-PCR, the attachment of sequencing adapters takes about 10 minutes.
Locus-specific protocols:
We also offer methods to enrich for loci of interest using the PCR Barcoding Kit (SQK-PBK004) and PCR Sequencing Kit (SQK-PSK004) where users tag their own specific amplicons with the same 5’ group before barcoding to simplify post-PCR processing. This is done via a four-primer PCR outlined in the Four-Primer PCR (SQK-PSK004 or SQK-PBK004) protocol with a recommended input of 30 ng DNA. Users add a 5’ tail to their own primer sequences and this acts as a priming site for a set of barcoded “outer” primers which are supplied in the PCR Barcoding Kit. These primers contain the 5’ tags which facilitate the ligase-free attachment of Rapid Sequencing Adapters.
Rapid PCR Barcoding Kit (SQK-RPB004):
The Rapid PCR Barcoding Kit (SQK-RPB004) is a fast and simple method of preparing barcoded libraries for low quantities of gDNA (1‒5 ng), with only ~15 minutes of hands-on preparation time. The transposase simultaneously cleaves the template molecules in each sample and attaches tags, which contain primer binding sites to the cleaved ends. The kit contains 12 primers which are then used to amplify each sample. Each primer contains a barcode and 5’ tag which facilitates the ligase-free attachment of Rapid Sequencing Adapters. Amplified and barcoded samples are then pooled together and Rapid Sequencing Adapters are added to the pooled mix.
16S Barcoding Kit (SQK-RAB204)
The 16S Barcoding Kit (SQK-RAB204) offers a method of amplifying and barcoding the ~1500 bp 16S rRNA gene from multiple samples and sequencing them together. By narrowing down to a specific region of interest, a user can see all the organisms in the sample without sequencing unnecessary regions of the genome, making the identification quicker and more economical. There are up to 12 unique barcodes, allowing the user to pool up to 12 different samples in one sequencing experiment using 10 ng of high molecular weight gDNA per sample as input. The DNA is amplified by PCR using the specific 16S barcode primers (27F and 1492R) and 5’ tags which facilitate the ligase-free attachment of Rapid Sequencing Adapters.
16S Barcoding Kit 1-24 (SQK-16S024):
The 16S Barcoding Kit 1-24 (SQK-16S024) offers a method of amplifying and barcoding the ~1500 bp 16S rRNA gene from multiple samples and sequencing them together. By narrowing down to a specific region of interest, a user can see all the organisms in the sample without sequencing unnecessary regions of the genome, making the identification quicker and more economical. There are up to 24 unique barcodes, allowing the user to pool up to 24 different samples in one sequencing experiment with 10 ng of high molecular weight gDNA per sample as input. The DNA is amplified by PCR using the specific 16S barcode primers (27F and 1492R) and 5’ tags which facilitate the ligase-free attachment of Rapid Sequencing Adapters.
cDNA barcoding kits
PCR-cDNA Barcoding Kit (SQK-PCB109)
This kit is the barcoding version of the cDNA-PCR Sequencing Kit (SQK-PCS109) and is recommended for users who want to multiplex cDNA samples with limited input material, whilst optimising for output. This kit may also be used to identify and quantify full-length transcripts or characterise and quantify isoforms, splice variants and fusion transcripts. This is also appropriate for users who are interested in differential gene expression or differential transcript usage. Up to 12 samples of cDNA can be prepared from an input as low as 1 ng of poly(A) RNA. Users who do not have poly(A)+ enriched RNA can use 50 ng of total RNA but additional optimisation may be required, as further explained below.
Complementary strand synthesis and strand switching are performed on full-length poly(A) RNA using the kit supplied oligonucleotides. There are 12 primer pairs that are used to generate and then amplify double-stranded cDNA by PCR amplification. Each primer pair contains a barcode and a 5’ tag which facilitates the ligase-free attachment of Rapid Sequencing Adapters. Amplified and barcoded samples are then pooled together and sequencing adapters are added to the pooled mix.
PCR-cDNA Barcoding Kit (SQK-PCB111.24):
This kit is the barcoding version of the cDNA-PCR Sequencing Kit (SQK-PCS111) and is recommended for users who want to multiplex cDNA samples with limited input material, whilst optimising for output. This kit may also be used to identify and quantify full-length transcripts or characterise and quantify isoforms, splice variants and fusion transcripts. This is also appropriate for users who are interested in differential gene expression or differential transcript usage. Up to 12 samples of cDNA can be prepared from an input as low as 1 ng of poly(A) RNA. Users who do not have poly(A)+ enriched RNA can use 50 ng of total RNA but additional optimisation may be required, as further explained below.
We have recently upgraded this kit to Kit 11 chemistry (SQK-PCB111.24) and increased the number of barcodes available to 24. This upgrade requires starting inputs of 4 ng poly(A) RNA or 200 ng of total RNA. Further the Kit 11 upgrades includes a new Rapid Adapter T (RAP T), which has a higher capture rate and fuel fix technology for longer experiments without the need for fuel addition during the run.
We have also included a new cDNA RT adapter (CRTA), Annealing Buffer (AB) and RT primer (RTP) to prime cDNA synthesis from the end of a transcript to reduce overlaps during the reverse transcription step. This also enables users to measure poly(A) tail lengths.
The kit workflow includes complementary strand synthesis and strand switching of full-length poly(A) RNA using the kit supplied oligonucleotides. There are 24 primer pairs that are used to generate and then amplify double-stranded cDNA by PCR amplification. Each primer pair contains a barcode and a 5’ tag which facilitates the ligase-free attachment of Rapid Sequencing Adapters. Amplified and barcoded samples are then pooled together and sequencing adapters are added to the pooled mix.
14. Barcode sequences
Native barcode sequences
Below is the full list of our native barcode (NB01-96) sequences. The first 24 unique barcodes are available in the Native Barcoding Kit 24 V14 (SQK-NBD114.24). The Native Barcoding Kit 96 V14 (SQK-NBD114.96) include the first 24 native barcodes, with the additional 72 unique barcodes. The native barcodes are shipped at 640 nM.
In addition to the barcodes, there are also flanking sequences which add an extra level of context during analysis.
Barcode flanking sequences:
Forward sequence: 5' - AAGGTTAA - barcode - CAGCACCT - 3' Reverse sequence: 5' - GGTGCTG - barcode - TTAACCTTAGCAAT - 3'
Native barcode sequences
| Component | Forward sequence | Reverse sequence |
|---|---|---|
| NB01 | CACAAAGACACCGACAACTTTCTT | AAGAAAGTTGTCGGTGTCTTTGTG |
| NB02 | ACAGACGACTACAAACGGAATCGA | TCGATTCCGTTTGTAGTCGTCTGT |
| NB03 | CCTGGTAACTGGGACACAAGACTC | GAGTCTTGTGTCCCAGTTACCAGG |
| NB04 | TAGGGAAACACGATAGAATCCGAA | TTCGGATTCTATCGTGTTTCCCTA |
| NB05 | AAGGTTACACAAACCCTGGACAAG | CTTGTCCAGGGTTTGTGTAACCTT |
| NB06 | GACTACTTTCTGCCTTTGCGAGAA | TTCTCGCAAAGGCAGAAAGTAGTC |
| NB07 | AAGGATTCATTCCCACGGTAACAC | GTGTTACCGTGGGAATGAATCCTT |
| NB08 | ACGTAACTTGGTTTGTTCCCTGAA | TTCAGGGAACAAACCAAGTTACGT |
| NB09 | AACCAAGACTCGCTGTGCCTAGTT | AACTAGGCACAGCGAGTCTTGGTT |
| NB10 | GAGAGGACAAAGGTTTCAACGCTT | AAGCGTTGAAACCTTTGTCCTCTC |
| NB11 | TCCATTCCCTCCGATAGATGAAAC | GTTTCATCTATCGGAGGGAATGGA |
| NB12 | TCCGATTCTGCTTCTTTCTACCTG | CAGGTAGAAAGAAGCAGAATCGGA |
| NB13 | AGAACGACTTCCATACTCGTGTGA | TCACACGAGTATGGAAGTCGTTCT |
| NB14 | AACGAGTCTCTTGGGACCCATAGA | TCTATGGGTCCCAAGAGACTCGTT |
| NB15 | AGGTCTACCTCGCTAACACCACTG | CAGTGGTGTTAGCGAGGTAGACCT |
| NB16 | CGTCAACTGACAGTGGTTCGTACT | AGTACGAACCACTGTCAGTTGACG |
| NB17 | ACCCTCCAGGAAAGTACCTCTGAT | ATCAGAGGTACTTTCCTGGAGGGT |
| NB18 | CCAAACCCAACAACCTAGATAGGC | GCCTATCTAGGTTGTTGGGTTTGG |
| NB19 | GTTCCTCGTGCAGTGTCAAGAGAT | ATCTCTTGACACTGCACGAGGAAC |
| NB20 | TTGCGTCCTGTTACGAGAACTCAT | ATGAGTTCTCGTAACAGGACGCAA |
| NB21 | GAGCCTCTCATTGTCCGTTCTCTA | TAGAGAACGGACAATGAGAGGCTC |
| NB22 | ACCACTGCCATGTATCAAAGTACG | CGTACTTTGATACATGGCAGTGGT |
| NB23 | CTTACTACCCAGTGAACCTCCTCG | CGAGGAGGTTCACTGGGTAGTAAG |
| NB24 | GCATAGTTCTGCATGATGGGTTAG | CTAACCCATCATGCAGAACTATGC |
| NB25 | GTAAGTTGGGTATGCAACGCAATG | CATTGCGTTGCATACCCAACTTAC |
| NB26 | CATACAGCGACTACGCATTCTCAT | ATGAGAATGCGTAGTCGCTGTATG |
| NB27 | CGACGGTTAGATTCACCTCTTACA | TGTAAGAGGTGAATCTAACCGTCG |
| NB28 | TGAAACCTAAGAAGGCACCGTATC | GATACGGTGCCTTCTTAGGTTTCA |
| NB29 | CTAGACACCTTGGGTTGACAGACC | GGTCTGTCAACCCAAGGTGTCTAG |
| NB30 | TCAGTGAGGATCTACTTCGACCCA | TGGGTCGAAGTAGATCCTCACTGA |
| NB31 | TGCGTACAGCAATCAGTTACATTG | CAATGTAACTGATTGCTGTACGCA |
| NB32 | CCAGTAGAAGTCCGACAACGTCAT | ATGACGTTGTCGGACTTCTACTGG |
| NB33 | CAGACTTGGTACGGTTGGGTAACT | AGTTACCCAACCGTACCAAGTCTG |
| NB34 | GGACGAAGAACTCAAGTCAAAGGC | GCCTTTGACTTGAGTTCTTCGTCC |
| NB35 | CTACTTACGAAGCTGAGGGACTGC | GCAGTCCCTCAGCTTCGTAAGTAG |
| NB36 | ATGTCCCAGTTAGAGGAGGAAACA | TGTTTCCTCCTCTAACTGGGACAT |
| NB37 | GCTTGCGATTGATGCTTAGTATCA | TGATACTAAGCATCAATCGCAAGC |
| NB38 | ACCACAGGAGGACGATACAGAGAA | TTCTCTGTATCGTCCTCCTGTGGT |
| NB39 | CCACAGTGTCAACTAGAGCCTCTC | GAGAGGCTCTAGTTGACACTGTGG |
| NB40 | TAGTTTGGATGACCAAGGATAGCC | GGCTATCCTTGGTCATCCAAACTA |
| NB41 | GGAGTTCGTCCAGAGAAGTACACG | CGTGTACTTCTCTGGACGAACTCC |
| NB42 | CTACGTGTAAGGCATACCTGCCAG | CTGGCAGGTATGCCTTACACGTAG |
| NB43 | CTTTCGTTGTTGACTCGACGGTAG | CTACCGTCGAGTCAACAACGAAAG |
| NB44 | AGTAGAAAGGGTTCCTTCCCACTC | GAGTGGGAAGGAACCCTTTCTACT |
| NB45 | GATCCAACAGAGATGCCTTCAGTG | CACTGAAGGCATCTCTGTTGGATC |
| NB46 | GCTGTGTTCCACTTCATTCTCCTG | CAGGAGAATGAAGTGGAACACAGC |
| NB47 | GTGCAACTTTCCCACAGGTAGTTC | GAACTACCTGTGGGAAAGTTGCAC |
| NB48 | CATCTGGAACGTGGTACACCTGTA | TACAGGTGTACCACGTTCCAGATG |
| NB49 | ACTGGTGCAGCTTTGAACATCTAG | CTAGATGTTCAAAGCTGCACCAGT |
| NB50 | ATGGACTTTGGTAACTTCCTGCGT | ACGCAGGAAGTTACCAAAGTCCAT |
| NB51 | GTTGAATGAGCCTACTGGGTCCTC | GAGGACCCAGTAGGCTCATTCAAC |
| NB52 | TGAGAGACAAGATTGTTCGTGGAC | GTCCACGAACAATCTTGTCTCTCA |
| NB53 | AGATTCAGACCGTCTCATGCAAAG | CTTTGCATGAGACGGTCTGAATCT |
| NB54 | CAAGAGCTTTGACTAAGGAGCATG | CATGCTCCTTAGTCAAAGCTCTTG |
| NB55 | TGGAAGATGAGACCCTGATCTACG | CGTAGATCAGGGTCTCATCTTCCA |
| NB56 | TCACTACTCAACAGGTGGCATGAA | TTCATGCCACCTGTTGAGTAGTGA |
| NB57 | GCTAGGTCAATCTCCTTCGGAAGT | ACTTCCGAAGGAGATTGACCTAGC |
| NB58 | CAGGTTACTCCTCCGTGAGTCTGA | TCAGACTCACGGAGGAGTAACCTG |
| NB59 | TCAATCAAGAAGGGAAAGCAAGGT | ACCTTGCTTTCCCTTCTTGATTGA |
| NB60 | CATGTTCAACCAAGGCTTCTATGG | CCATAGAAGCCTTGGTTGAACATG |
| NB61 | AGAGGGTACTATGTGCCTCAGCAC | GTGCTGAGGCACATAGTACCCTCT |
| NB62 | CACCCACACTTACTTCAGGACGTA | TACGTCCTGAAGTAAGTGTGGGTG |
| NB63 | TTCTGAAGTTCCTGGGTCTTGAAC | GTTCAAGACCCAGGAACTTCAGAA |
| NB64 | GACAGACACCGTTCATCGACTTTC | GAAAGTCGATGAACGGTGTCTGTC |
| NB65 | TTCTCAGTCTTCCTCCAGACAAGG | CCTTGTCTGGAGGAAGACTGAGAA |
| NB66 | CCGATCCTTGTGGCTTCTAACTTC | GAAGTTAGAAGCCACAAGGATCGG |
| NB67 | GTTTGTCATACTCGTGTGCTCACC | GGTGAGCACACGAGTATGACAAAC |
| NB68 | GAATCTAAGCAAACACGAAGGTGG | CCACCTTCGTGTTTGCTTAGATTC |
| NB69 | TACAGTCCGAGCCTCATGTGATCT | AGATCACATGAGGCTCGGACTGTA |
| NB70 | ACCGAGATCCTACGAATGGAGTGT | ACACTCCATTCGTAGGATCTCGGT |
| NB71 | CCTGGGAGCATCAGGTAGTAACAG | CTGTTACTACCTGATGCTCCCAGG |
| NB72 | TAGCTGACTGTCTTCCATACCGAC | GTCGGTATGGAAGACAGTCAGCTA |
| NB73 | AAGAAACAGGATGACAGAACCCTC | GAGGGTTCTGTCATCCTGTTTCTT |
| NB74 | TACAAGCATCCCAACACTTCCACT | AGTGGAAGTGTTGGGATGCTTGTA |
| NB75 | GACCATTGTGATGAACCCTGTTGT | ACAACAGGGTTCATCACAATGGTC |
| NB76 | ATGCTTGTTACATCAACCCTGGAC | GTCCAGGGTTGATGTAACAAGCAT |
| NB77 | CGACCTGTTTCTCAGGGATACAAC | GTTGTATCCCTGAGAAACAGGTCG |
| NB78 | AACAACCGAACCTTTGAATCAGAA | TTCTGATTCAAAGGTTCGGTTGTT |
| NB79 | TCTCGGAGATAGTTCTCACTGCTG | CAGCAGTGAGAACTATCTCCGAGA |
| NB80 | CGGATGAACATAGGATAGCGATTC | GAATCGCTATCCTATGTTCATCCG |
| NB81 | CCTCATCTTGTGAAGTTGTTTCGG | CCGAAACAACTTCACAAGATGAGG |
| NB82 | ACGGTATGTCGAGTTCCAGGACTA | TAGTCCTGGAACTCGACATACCGT |
| NB83 | TGGCTTGATCTAGGTAAGGTCGAA | TTCGACCTTACCTAGATCAAGCCA |
| NB84 | GTAGTGGACCTAGAACCTGTGCCA | TGGCACAGGTTCTAGGTCCACTAC |
| NB85 | AACGGAGGAGTTAGTTGGATGATC | GATCATCCAACTAACTCCTCCGTT |
| NB86 | AGGTGATCCCAACAAGCGTAAGTA | TACTTACGCTTGTTGGGATCACCT |
| NB87 | TACATGCTCCTGTTGTTAGGGAGG | CCTCCCTAACAACAGGAGCATGTA |
| NB88 | TCTTCTACTACCGATCCGAAGCAG | CTGCTTCGGATCGGTAGTAGAAGA |
| NB89 | ACAGCATCAATGTTTGGCTAGTTG | CAACTAGCCAAACATTGATGCTGT |
| NB90 | GATGTAGAGGGTACGGTTTGAGGC | GCCTCAAACCGTACCCTCTACATC |
| NB91 | GGCTCCATAGGAACTCACGCTACT | AGTAGCGTGAGTTCCTATGGAGCC |
| NB92 | TTGTGAGTGGAAAGATACAGGACC | GGTCCTGTATCTTTCCACTCACAA |
| NB93 | AGTTTCCATCACTTCAGACTTGGG | CCCAAGTCTGAAGTGATGGAAACT |
| NB94 | GATTGTCCTCAAACTGCCACCTAC | GTAGGTGGCAGTTTGAGGACAATC |
| NB95 | CCTGTCTGGAAGAAGAATGGACTT | AAGTCCATTCTTCTTCCAGACAGG |
| NB96 | CTGAACGGTCATAGAGTCCACCAT | ATGGTGGACTCTATGACCGTTCAG |
Rapid barcode sequences
Below is a full list of the rapid barcode sequences used across the rapid-based barcoding kits, with the component acronym for the specific sequencing kit.
Note: These are not the full sequences due to proprietary information.
In addition to the barcodes, there are also flanking sequences which add an extra level of context during analysis.
Rapid barcoding kits
Rapid Barcoding Kit 24 V14 (SQK-RBK114.24): RB01-24
Rapid Barcoding Kit 96 V14 (SQK-RBK114.96): RB01-96
Legacy kits
Rapid Barcoding Kit (SQK-RBK004): RB01-12
Rapid Barcoding Kit 96 (SQK-RBK110.96): RB01-96
Barcode flanking sequence:
5' - GCTTGGGTGTTTAACC - barcode - GTTTTCGCATTTATCGTGAAACGCTTTCGCGTTTTTCGTGCGCCGCTTCA - 3'
PCR barcoding kits
PCR-cDNA Barcoding Kit 24 V14 (SQK-PCB114.24): BP01-24
Legacy kits
PCR-cDNA Barcoding Kit 24 (SQK-PCB111.24): BP01-24
PCR-cDNA Barcoding Kit (SQK-PCB109): BP01-12
PCR Barcoding Kit (SQK-PBK004): BP01-12
Barcode flanking sequence:
Top strand: 5' - ATCGCCTACCGTGA - barcode - TTGCCTGTCGCTCTATCTTC - 3'
Bottom strand: 5' - ATCGCCTACCGTGA - barcode - TCTGTTGGTGCTGATATTGC - 3'
The top and bottom barcode flanking sequences are different to avoid 5' and 3' end sequences annealing to each other and forming a loop.
16S barcoding kits
16S Barcoding Kit 24 V14 (SQK-16S114.24): 16S01-24
Legacy kits
16S Barcoding Kit 1-24 (SQK-16S024): 16S01-24
16S Barcoding Kit (SQK-RAB204): 16S01-12
Barcode flanking sequence:
Forward 16S primer: 5' - ATCGCCTACCGTGAC - barcode - AGAGTTTGATCMTGGCTCAG - 3'
Reverse 16S primer: 5' - ATCGCCTACCGTGAC - barcode - CGGTTACCTTGTTACGACTT - 3'
The 3' flanking sequence of the forward primer contains a wobble base (denoted by M; in the primer the base is either an A or a C) in a variable region of the 16S gene.
Rapid PCR barcoding kits
Rapid PCR Barcoding Kit 24 V14 (SQK-RPB114.24): RLB01-24
Legacy kit
Rapid PCR Barcoding Kit (SQK-RPB004): RLB01-12A
Barcode flanking sequence:
5' - ATCGCCTACCGTGAC - barcode - CGTTTTTCGTGCGCCGCTTC - 3'
PCR barcoding expansions
PCR Barcoding Expansion 1-12 (EXP-PBC001): BC01-12
PCR Barcoding Expansion 1-96 (EXP-PBC096): BC01-96
Barcode flanking sequence:
5' - GGTGCTG - barcode - TTAACCT - 3'
| Component | Sequence |
|---|---|
| BP01 / BC01 / RB01 / 16S01 / RLB01 | AAGAAAGTTGTCGGTGTCTTTGTG |
| BP02 / BC02 / RB02 / 16S02 / RLB02 | TCGATTCCGTTTGTAGTCGTCTGT |
| BP03 / BC03 / RB03 / 16S03 / RLB03 | GAGTCTTGTGTCCCAGTTACCAGG |
| BP04 / BC04 / RB04 / 16S04 / RLB04 | TTCGGATTCTATCGTGTTTCCCTA |
| BP05 / BC05 / RB05 / 16S05 / RLB05 | CTTGTCCAGGGTTTGTGTAACCTT |
| BP06 / BC06 / RB06 / 16S06 / RLB06 | TTCTCGCAAAGGCAGAAAGTAGTC |
| BP07 / BC07 / RB07 / 16S07 / RLB07 | GTGTTACCGTGGGAATGAATCCTT |
| BP08 / BC08 / RB08 / 16S08 / RLB08 | TTCAGGGAACAAACCAAGTTACGT |
| BP09 / BC09 / RB09 / 16S09 / RLB09 | AACTAGGCACAGCGAGTCTTGGTT |
| BP10 / BC10 / RB10 / 16S10 / RLB10 | AAGCGTTGAAACCTTTGTCCTCTC |
| BP11 / BC11 / RB11 / 16S11 / RLB11 | GTTTCATCTATCGGAGGGAATGGA |
| BP12 / BC12 / RB12 / 16S12 | CAGGTAGAAAGAAGCAGAATCGGA |
| RLB12 (in current chemistry kits V14+ / RLB012A in legacy kits) | GTTGAGTTACAAAGCACCGATCAG |
| BP13 / BC13 / RB13 / 16S13 / RLB13 | AGAACGACTTCCATACTCGTGTGA |
| BP14 / BC14 / RB14 / 16S14 / RLB14 | AACGAGTCTCTTGGGACCCATAGA |
| BP15 / BC15 / RB15 / 16S15 / RLB15 | AGGTCTACCTCGCTAACACCACTG |
| BP16 / BC16 / RB16 / 16S16 / RLB16 | CGTCAACTGACAGTGGTTCGTACT |
| BP17 / BC17 / RB17 / 16S17 / RLB17 | ACCCTCCAGGAAAGTACCTCTGAT |
| BP18 / BC18 / RB18 / 16S18 / RLB18 | CCAAACCCAACAACCTAGATAGGC |
| BP19 / BC19 / RB19 / 16S19 / RLB19 | GTTCCTCGTGCAGTGTCAAGAGAT |
| BP20 / BC20 / RB20 / 16S20 / RLB20 | TTGCGTCCTGTTACGAGAACTCAT |
| BP21 / BC21 / RB21 / 16S21 / RLB21 | GAGCCTCTCATTGTCCGTTCTCTA |
| BP22 / BC22 / RB22 / 16S22 / RLB22 | ACCACTGCCATGTATCAAAGTACG |
| BP23 / BC23 / RB23 / 16S23 / RLB23 | CTTACTACCCAGTGAACCTCCTCG |
| BP24 / BC24 / RB24 / 16S24 / RLB24 | GCATAGTTCTGCATGATGGGTTAG |
| BC25 / RB25 | GTAAGTTGGGTATGCAACGCAATG |
| BC26 / RB26 | CATACAGCGACTACGCATTCTCAT |
| BC27 / RB27 | CGACGGTTAGATTCACCTCTTACA |
| BC28 / RB28 | TGAAACCTAAGAAGGCACCGTATC |
| BC29 / RB29 | CTAGACACCTTGGGTTGACAGACC |
| BC30 / RB30 | TCAGTGAGGATCTACTTCGACCCA |
| BC31 / RB31 | TGCGTACAGCAATCAGTTACATTG |
| BC32 / RB32 | CCAGTAGAAGTCCGACAACGTCAT |
| BC33 / RB33 | CAGACTTGGTACGGTTGGGTAACT |
| BC34 / RB34 | GGACGAAGAACTCAAGTCAAAGGC |
| BC35 / RB35 | CTACTTACGAAGCTGAGGGACTGC |
| BC36 / RB36 | ATGTCCCAGTTAGAGGAGGAAACA |
| BC37 / RB37 | GCTTGCGATTGATGCTTAGTATCA |
| BC38 / RB38 | ACCACAGGAGGACGATACAGAGAA |
| BC39 / RB39 | CCACAGTGTCAACTAGAGCCTCTC |
| BC40 / RB40 | TAGTTTGGATGACCAAGGATAGCC |
| BC41 / RB41 | GGAGTTCGTCCAGAGAAGTACACG |
| BC42 / RB42 | CTACGTGTAAGGCATACCTGCCAG |
| BC43 / RB43 | CTTTCGTTGTTGACTCGACGGTAG |
| BC44 / RB44 | AGTAGAAAGGGTTCCTTCCCACTC |
| BC45 / RB45 | GATCCAACAGAGATGCCTTCAGTG |
| BC46 / RB46 | GCTGTGTTCCACTTCATTCTCCTG |
| BC47 / RB47 | GTGCAACTTTCCCACAGGTAGTTC |
| BC48 / RB48 | CATCTGGAACGTGGTACACCTGTA |
| BC49 / RB49 | ACTGGTGCAGCTTTGAACATCTAG |
| BC50 / RB50 | ATGGACTTTGGTAACTTCCTGCGT |
| BC51 / RB51 | GTTGAATGAGCCTACTGGGTCCTC |
| BC52 / RB52 | TGAGAGACAAGATTGTTCGTGGAC |
| BC53 / RB53 | AGATTCAGACCGTCTCATGCAAAG |
| BC54 / RB54 | CAAGAGCTTTGACTAAGGAGCATG |
| BC55 / RB55 | TGGAAGATGAGACCCTGATCTACG |
| BC56 / RB56 | TCACTACTCAACAGGTGGCATGAA |
| BC57 / RB57 | GCTAGGTCAATCTCCTTCGGAAGT |
| BC58 / RB58 | CAGGTTACTCCTCCGTGAGTCTGA |
| BC59 / RB59 | TCAATCAAGAAGGGAAAGCAAGGT |
| BC60 / RB60 | CATGTTCAACCAAGGCTTCTATGG |
| BC61 / RB61 | AGAGGGTACTATGTGCCTCAGCAC |
| BC62 / RB62 | CACCCACACTTACTTCAGGACGTA |
| BC63 / RB63 | TTCTGAAGTTCCTGGGTCTTGAAC |
| BC64 / RB64 | GACAGACACCGTTCATCGACTTTC |
| BC65 / RB65 | TTCTCAGTCTTCCTCCAGACAAGG |
| BC66 / RB66 | CCGATCCTTGTGGCTTCTAACTTC |
| BC67 / RB67 | GTTTGTCATACTCGTGTGCTCACC |
| BC68 / RB68 | GAATCTAAGCAAACACGAAGGTGG |
| BC69 / RB69 | TACAGTCCGAGCCTCATGTGATCT |
| BC70 / RB70 | ACCGAGATCCTACGAATGGAGTGT |
| BC71 / RB71 | CCTGGGAGCATCAGGTAGTAACAG |
| BC72 / RB72 | TAGCTGACTGTCTTCCATACCGAC |
| BC73 / RB73 | AAGAAACAGGATGACAGAACCCTC |
| BC74 / RB74 | TACAAGCATCCCAACACTTCCACT |
| BC75 / RB75 | GACCATTGTGATGAACCCTGTTGT |
| BC76 / RB76 | ATGCTTGTTACATCAACCCTGGAC |
| BC77 / RB77 | CGACCTGTTTCTCAGGGATACAAC |
| BC78 / RB78 | AACAACCGAACCTTTGAATCAGAA |
| BC79 / RB79 | TCTCGGAGATAGTTCTCACTGCTG |
| BC80 / RB80 | CGGATGAACATAGGATAGCGATTC |
| BC81 / RB81 | CCTCATCTTGTGAAGTTGTTTCGG |
| BC82 / RB82 | ACGGTATGTCGAGTTCCAGGACTA |
| BC83 / RB83 | TGGCTTGATCTAGGTAAGGTCGAA |
| BC84 / RB84 | GTAGTGGACCTAGAACCTGTGCCA |
| BC85 / RB85 | AACGGAGGAGTTAGTTGGATGATC |
| BC86 / RB86 | AGGTGATCCCAACAAGCGTAAGTA |
| BC87 / RB87 | TACATGCTCCTGTTGTTAGGGAGG |
| BC88 / RB88 | TCTTCTACTACCGATCCGAAGCAG |
| BC89 / RB89 | ACAGCATCAATGTTTGGCTAGTTG |
| BC90 / RB90 | GATGTAGAGGGTACGGTTTGAGGC |
| BC91 / RB91 | GGCTCCATAGGAACTCACGCTACT |
| BC92 / RB92 | TTGTGAGTGGAAAGATACAGGACC |
| BC93 / RB93 | AGTTTCCATCACTTCAGACTTGGG |
| BC94 / RB94 | GATTGTCCTCAAACTGCCACCTAC |
| BC95 / RB95 | CCTGTCTGGAAGAAGAATGGACTT |
| BC96 / RB96 | CTGAACGGTCATAGAGTCCACCAT |
Amplicon barcode sequences
Below is a full list of the amplicon barcode sequences used across the amplicon barcoding kits, with the component acronym for the specific sequencing kit.
Note: These are not the full sequences due to proprietory information.
In addition to the barcodes, there are also flanking sequences which add an extra level of context during analysis.
Amplicon barcoding kits
Microbial Amplicon Barcoding Kit 24 V14 (SQK-MAB114.24): AB01-24
Barcode flanking sequence:
5' - GCTTGGGTGTTTAACC - barcode - CCATATCCGTGTCGCCCTTATTCCGATAGTGACTACAGCGGCATGAGTG - 3'
| Component | Sequence |
|---|---|
| AB01 | GCACCTGGAACTTGTGCCTTCCAC |
| AB02 | CCGAAATAGGTTATCTGTTGTTGT |
| AB03 | ATCAATCGCTGGACGATGGATTAG |
| AB04 | CCACCCGCTCCTGCCGGTGGGCGT |
| AB05 | AGACTCTTGGGCTCGCCACGTCCC |
| AB06 | TCTGTATCCGGAGACGGGATGGAC |
| AB07 | TTTCGGATCAATCGACCGCAAACG |
| AB08 | ACTCAAACATTCTGTTAGATCGCG |
| AB09 | AAATGGAACCCGGATATGTTTACT |
| AB10 | TAAATCGACCTATGATGAACACAG |
| AB11 | ACATGTTGGAGTGAAAGTCGGGTA |
| AB12 | CCTGGACCACGATCATTGTAACAT |
| AB13 | TATGGTGGATCTCCCTCTATCTTC |
| AB14 | AAGTAAATGGGACGCCCACTCCGA |
| AB15 | TGTTCGCGGCTTGATCTAATATTA |
| AB16 | AGAGAGCTTCCCGGGAGGGTGGTC |
| AB17 | TTGTGAATATCTGTCACAAACACC |
| AB18 | CAATCGTACCAGGGAACATAAAGT |
| AB19 | CACACCCAAACAATATGGACCCGT |
| AB20 | AATAACCACATCCGCCCTCCGCAC |
| AB21 | TCCTAATAATGTGTAGATCGGTCC |
| AB22 | AGTCGATGGAACAAGAGAAGTTAT |
| AB23 | AAACTCACTGTATGTCGTTTCTAT |
| AB24 | TGACATCACTGATCGAGGAAGATC |
15. Adapter sequences
Sequencing adapter sequences
Below are the adapter sequences for our current kits.
Ligation Adapter (LA)
Top strand: 5'-TTTTTTTTCCTGTACTTCGTTCAGTTACGTATTGCT-3’
Bottom strand: 5’-GCAATACGTAACTGAACGAAGTACAGG-3’
Native Adapter (NA)
Top strand: 5'-TTTTTTTTCCTGTACTTCGTTCAGTTACGTATTGCT-3'
Bottom strand: 5'-ACGTAACTGAACGAAGTACAGG-3'
Rapid Adapter (RA)
Top strand: 5’-TTTTTTTTCCTGTACTTCGTTCAGTTACGTATTGCT-3’
RT Primer (RTP)
5’-CTTGCCTGTCGCTCTATCTTCAGAGGAG-3’
Strand Switching Primer II (SSPII)
5’-TTTCTGTTGGTGCTGATATTGCTTTVVVVTTVVVVTTVVVVTTVVVVTTTmGmGmG-3’
cDNA RT Adapter (CRTA)
5'-CTTGCGGGCGGCGGACTCTCCTCTGAAGATAGAGCGACAGGCAAG-3'
Rapid Adapter T (RAP T)
5'-TTTTTTTTCCTGTACTTCGTTCAGTTACGTATTGCT-3’
cDNA Primer (cPRM)
Forward sequence: 5'-ATCGCCTACCGTGACAAGAAAGTTGTCGGTGTCTTTGTGACTTGCCTGTCGCTCTATCTTC-3’
Reverse sequence: 5’-ATCGCCTACCGTGACAAGAAAGTTGTCGGTGTCTTTGTGTTTCTGTTGGTGCTGATATTGC-3’
16. Kit reagents
Sequencing kit reagents
Below is a list of the common reagents included in our sequencing kits.
Ligation Adapter (LA) The sequencing adapter is ligated onto the dA-tailed DNA fragments during library preparation. A motor protein is loaded onto the sequencing adapter and regulates the DNA passage through the nanopore.
Native Adapter (NA) The sequencing adapter ligates to the native barcodes ligated onto the dA-tailed DNA fragments during the library preparation of the native barcoding kits. A motor protein is loaded onto the sequencing adapter and regulates the DNA passage through the nanopore.
Fragmentation Mix (FRA) This reagent contains a transposase to fragment DNA strands during library preparation and simultaneously attaches transposase adapters to the DNA ends.
Rapid Adapter (RA) The sequencing adapter is attached to the transposase adapters in the Fragmentation Mix (FRA). A motor protein is loaded onto the sequencing adapter and regulates the DNA passage through the nanopore.
Adapter Buffer (ADB) This buffer is used in conjunction with the Rapid Adapter (RA) during the adapter attachment step to reach the correct concentration for optimal adaptation.
Native Barcodes (NB01-96) Barcodes are a known sequence used to identify samples in a pooled library. The Native Barcodes are specific to this kit and are ligated onto the DNA ends after end-prep. Native barcodes are shipped at 640 nM.
EDTA (EDTA) This reagent is used during the barcoding step of the native barcoding kits to stop the barcode ligation reaction after incubation.
AMPure XP Beads (AXP) These beads are manufactured by Beckman Coulter, Inc. and are used in the clean up steps of library preparations to purify a reaction e.g. excess barcodes or transposases present in a reaction.
Sequencing Buffer (SB) The sequencing buffer provides fuel and the optimal chemical conditions for powering DNA translocation through the nanopore. In SQK-LSK114, this component has been upgraded to improve sequencing by reducing variability in current levels during experiment runs.
Short and Long Fragment Buffers Short Fragment Buffer (SFB) is used to enrich fragments of all sizes after adapter ligation. Long Fragment Buffer (LFB) is used to enrich fragments of ≥3 kb after adapter ligation.
Ligation Buffer (LNB) This buffer is used to aid ligation of the sequencing adapters.
Elution Buffer (EB) This buffer is optimised to keep the integrity of the motor protein on the Y-shaped adapter.
Library Beads (LIB) These beads help reduce frequency of blocking.
Library Solution (LIS) This reagent is the same solution that the Library Beads are stored in and are used to ease the loading of viscous libraries.
Flow Cell Flush (FCF) and Flow Cell Tether (FCT) These reagents are combined to make up the flow cell priming mix. The tether is used to bring the DNA strand to the membrane and to improve DNA capture by approximately 10,000-fold compared with capture without tether.
DNA Control Sample (DCS) The DNA Control Sample (DCS) is a 3.6 kb amplicon of the Lambda phage genome. The control sample is included in the library and added to DNA samples during DNA repair and end-prep for troubleshooting purposes.
Lambda DNA (LMD) The Lambda DNA (LMD) is a pure, high-quality 48.5 kb DNA (consisting of a complete genome of Enterobacteria phage lambda) that can be used to prepare a library and run a control experiment. The lambda control protocols allows a new user to practise the process of library preparation, and can also be used to troubleshoot poorly performing experiments.






