Secuenciación múltiple de metilación con representación reducida (RRMS) a partir de células mediante el kit SQK-NBD114.24 (RRMS_9209_v114_revI_21Oct2025)
PromethION: Protocol
Secuenciación múltiple de metilación con representación reducida (RRMS) a partir de células mediante el kit SQK-NBD114.24 V RRMS_9209_v114_revI_21Oct2025
Para uso exclusivo en investigación
FOR RESEARCH USE ONLY
Contents
Introducción al protocolo
Preparación de muestras
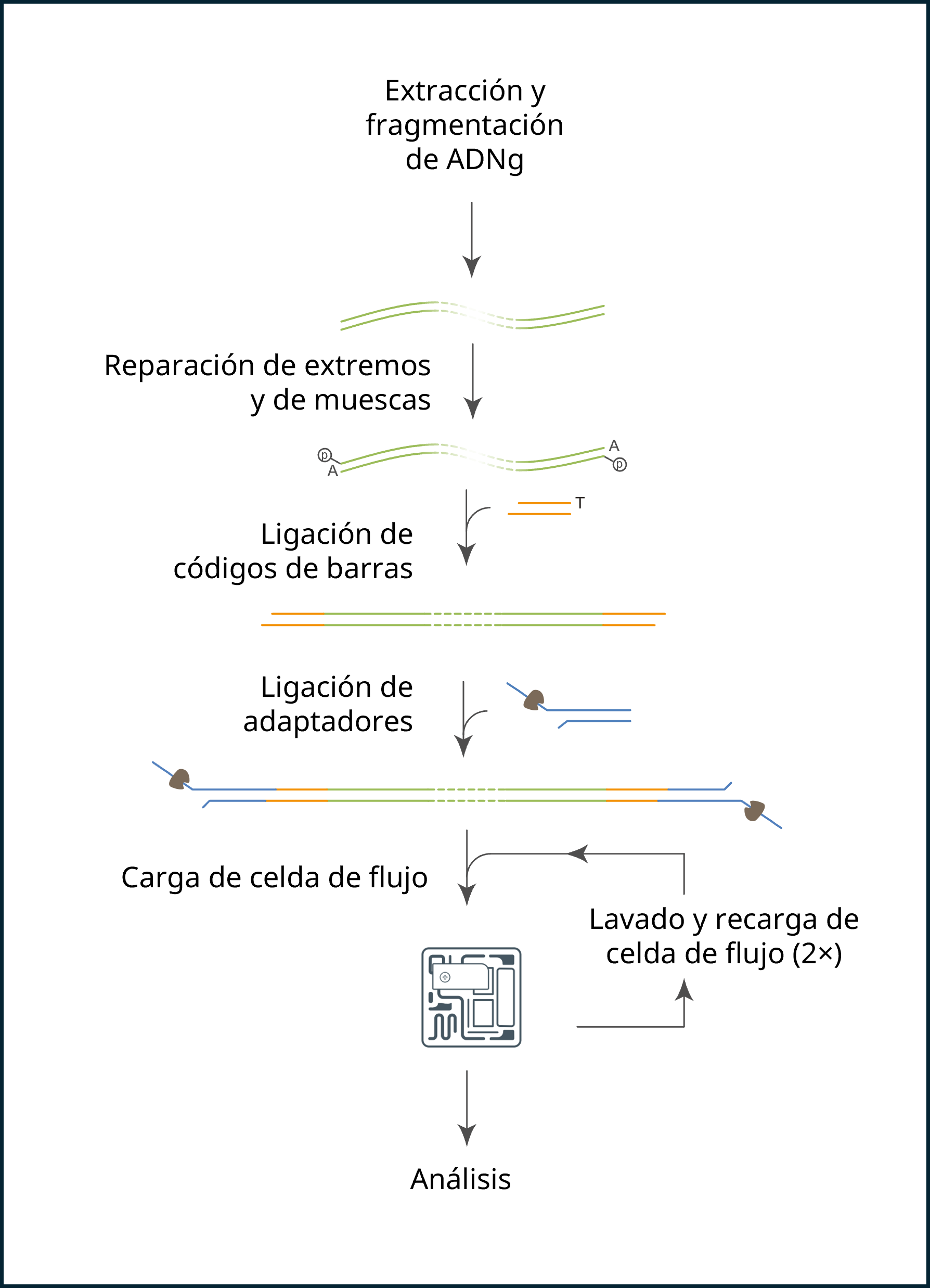
Preparación de la biblioteca
- 6. Reparación del ADN y preparación de los extremos
- 7. Ligación de los códigos de barras sin amplificación
- 8. Ligación de los adaptadores y purificación
- 9. Acondicionamiento y carga de la celda de flujo PromethION.
- 10. Lavado y recarga de celdas de flujo PromethION
Secuenciación y análisis de datos
- 11. Adquisición de datos e identificación de bases
- 12. Análisis de datos
- 13. Reutilización y retorno de las celdas de flujo
Resolución de problemas
Descripción general
Para uso exclusivo en investigación
1. Descripción general
Muestreo adaptativo en la química del kit 14
Aunque utilice química del kit 14, este flujo de trabajo se ha optimizado para enriquecer regiones específicas de interés (ROI) con muestreo adaptativo. De este modo, se garantiza un mayor rendimiento y mejores resultados de secuenciación.
Encontrará información básica sobre el diseño de experimentos con muestreo adaptativo en el documento técnico Buenas prácticas de muestreo adaptativo
Secuenciación múltiple de metilación con representación reducida (RRMS)
La secuenciación por nanoporos permite la detección directa de citosinas metiladas (p. Ej., en puntos CpG), sin necesidad de convertirlas en bisulfito. Los sitios CpG se dan con frecuencia en agrupaciones de elevada densidad llamadas islas CpG (CGI) y la mayoría de genes de vertebrados tienen sus promotores alojados dentro de estas islas.
Los cambios en los patrones de metilación dentro de los promotores están asociados a los cambios en la expresión génica y a los estados de enfermedades como el cáncer: analizar las diferencias de metilación entre muestras de tumores y muestras normales ayuda a descubrir mecanismos asociados con la formación y desarrollo de tumores.
El muestreo adaptativo (MA) constituye un método rápido, flexible y preciso de enriquecer regiones de interés (p. Ej., islas CpG) mediante el agotamiento de regiones excluidas durante la secuenciación, sin necesidad de manipular la muestra de antemano.
Encontrará más información sobre cómo funciona el método y cómo se compara con otras técnicas de análisis de metilación (p. ej., matrices EPIC, bisulfito), en el documento «Introducción a la secuenciación de metilación con representación reducida».
El protocolo de secuenciación múltiple de metilación con representación reducida es compatible con los dispositivos GridION, PromethION 2 Solo, P24 y P48.
Cuando se ejecute en GridION, recomendamos secuenciar una muestra por celda de flujo y utilizar nuestro protocolo Secuenciación de metilación con representación reducida (RRMS) a partir de células que utilizan el protocolo SQK-LSK114.
Secuenciación de muestras humanas
El protocolo RRMS permite dirigirse a 310 Mb del genoma humano, altamente enriquecidos en las regiones CpG, e incluye todas las islas CpG anotadas, sus zonas adyacentes (shores), regiones más alejadas (shelves) y más del 90 % de las regiones promotoras (el 100 % de los promotores que contienen más de 4 sitios CpG). También abarca otras regiones del genoma ricas en CpG. El número total de sitios CpG en el archivo BED es 7,18 millones.
A efectos de evaluación comparativa, hemos realizado un protocolo RRMS en cinco duplicados de una estirpe celular de un melanoma metastásico y su par normal para un individuo masculino (COLO829/COLO829_BL) y un par de estirpes celulares de cáncer de mama triple negativo (HCC1395/HCC1935_BL). Cada muestra se secuenció en una única celda de flujo MinION/GridION. El protocolo RRMS dio como resultado identificaciones de metilación de elevada confianza (>10 lecturas solapadas) en un rango de 7,3 a 8,5 millones de sitios CpG por muestra.
A modo de comparación, también realizamos el protocolo Secuenciación por bisulfito de representación reducida (RRBS), que por lo general produce entre 1,7 y 2,5 identificaciones de metilación con elevada confianza por muestra. Encontrará más información acerca de esta comparación en el documento sobre las prestaciones de RRMS y el póster.
Secuenciación de muestras de ratón
También se han desarrollado un protocolo RRMS y un nuevo archivo BED dirigido a 308 Mb del genoma del ratón, con una cobertura del 100 % de las islas CpG y las regiones promotoras, así como de otras regiones del genoma ricas en sitios CpG.
El funcionamiento de RRMS en muestras de ratón se caracterizó por el uso de réplicas de una estirpe de citoblastos embrionarios derivada de blastocitos, (ES-E14TG2a) y una estirpe celular de leucemia ((BALB/c AMuLV A.3R.1). También se secuenció, como control, una biblioteca que no era RRMS. Cada muestra se secuenció en una celda de flujo MinION: RRMS dio como resultado identificaciones de metilación de elevada fiabilidad (más de 10× lecturas por sitio) en 5,0-5,8 millones de sitios CpG por muestra en el genoma del ratón, comparado con ~400 000 sitios CpG en la biblioteca de control.
Con el protocolo RRMS y un archivo BED personalizado, es posible secuenciar otros genomas de vertebrados, pero Oxford Nanopore Technologies ha validado este método únicamente en muestras humanas y de ratón.
Introducción a la extracción de ADN y a la secuenciación múltiple con el protocolo RRMS
Este protocolo describe cómo llevar a cabo la extracción de ADN y secuenciación múltiple de metilación con representación reducida (RRMS) de hasta 4 muestras en una única celda de flujo PromethION, utilizando el kit Native Barcoding (SQK-NBD114.24) y la función Muestreo adaptativo de MinKNOW.
Pasos del proceso de secuenciación:
Preparación del experimento
Pasos:
- Tener a disposición el kit de secuenciación, el instrumental adecuado y los reactivos de otros fabricantes.
- Descargar el programa con el que se obtienen y analizan los datos.
- Contar con el archivo BED adecuado para realizar el muestreo adaptativo.
- Verificar la celda de flujo y comprobar que tiene poros suficientes para realizar una buena secuenciación.
Preparación de la muestra
- Extraer el ADN con ayuda del kit Puregene Cell, de QIAGEN.
- Fragmentar el ADN con ayuda del tubo g-TUBE de Covaris, comprobar su longitud, cantidad y pureza. Los controles de calidad realizados durante el protocolo son esenciales para garantizar el éxito del experimento.
Preparación de la biblioteca
La tabla siguiente describe los pasos necesarios en la preparación de la biblioteca.
| Preparación de la biblioteca | Proceso | Tiempo | Parada opcional |
|---|---|---|---|
| Reparación del ADN y preparación de los extremos | Reparar el ADN fragmentado y preparar los extremos de ADN para ligar los códigos de barras | 35 minutos | 4 °C hasta el día siguiente |
| Ligación de códigos de barras | Ligar los códigos de barras a los extremos de ADN | 60 minutos | 4 °C hasta el día siguiente |
| Ligación de los adaptadores y purificación | Ligar los adaptadores a los códigos de barras de los extremos de ADN | 50 minutos | Almacenamiento a 4 °C en usos a corto plazo o reutilización, como la recarga de la celda de flujo Almacenamiento a –80 °C cuando se trata de un solo uso o conservación a largo plazo. Recomendamos secuenciar la biblioteca tan pronto como se liguen los adaptadores. |
| Acondicionar y cargar la celda de flujo | Acondicionar la celda de flujo y cargar la biblioteca preparada | 10 minutos | |
| Lavar y volver a cargar las celdas de flujo (2×) | Detener la secuenciación. Lavar la celda de flujo con agua sin nucleasas para eliminar la carga de biblioteca anterior y desbloquear los poros. Acondicionar la celda de flujo y cargar la biblioteca | 60 minutos (2×) |
Secuenciación y análisis
Pasos:
- Empezar el proceso de secuenciación usando el programa MinKNOW, que recopilará los datos sin procesar del dispositivo y los convertirá en lecturas de bases identificadas. Mientras configura el experimento, active la función Muestreo adaptativo e importe un archivo BED preconfigurado con sus regiones de interés, junto a un archivo de referencia FASTA.
- Secuenciar la muestra durante un total de 96 horas, con dos lavados de celda de flujo cuando el recuento de poros disponible disminuya a un 40 % del recuento de poros inicial (por lo general, tras 24 horas aprox. y una segunda vez tras 48 horas).
- Utilizar Dorado para identificar bases modificadas; encontrará más información en la página de Dorado en github.
- Utilizar los comandos recomendados al final de este protocolo para agregar las bases modificadas y realizar anotaciones de islas CpG.
Compatibilidad del protocolo
Este protocolo debería usarse solo en combinación con los siguientes productos:
- Native Barcoding Kit 24 V14 (SQK-NBD114.24)
- Celdas de flujo PromethION R10.4.1 (FLO-PRO114M)
- Kit de lavado Flow Cell Wash Kit (EXP-WSH004)
- Sequencing Auxiliary Vials V14 (EXP-AUX003)
- Native Barcoding Expansion V14 (EXP-NBA114)
- Dispositivo PromethION 24/48 - Requisitos informáticos PromethION
- Dispositivo PromethION 2 Solo - Requisitos informáticos PromethION 2 Solo
2. Material y consumibles
Material
- 5 x 10^6 de células por muestra (EXTRACCIÓN)
- 2 µg de ADNg fragmentado por muestra (PREPARACIÓN DE LA BIBLIOTECA)
- Kit Native Barcoding 24 V14 (SQK-NBD114.24)
- Kit Flow Cell Wash (EXP-WSH004)
Consumibles
- Celdas de flujo PromethION
- Kit Puregene Cell (QIAGEN, 158043)
- Tampón TE (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) (Fisher Scientific, 10224683)
- PBS 1× (solución salina tamponada con fosfato)
- Isopropanol
- Tubo g-TUBE™ (Covaris, 520079)
- NEBNext FFPE Repair Mix (NEB M6630)
- NEBNext Ultra II End Repair/dA-tailing Module (NEB E7546)
- NEB Blunt/TA Ligase Master Mix (NEB, M0367)
- NEBNext® Quick Ligation Module (NEB, E6056)
- Etanol al 70 % recién preparado en agua sin nucleasas
- Etanol al 80 % recién preparado con agua sin nucleasas
- Nuclease-free water (e.g. Thermo Scientific, AM9937)
- Tubos Falcon de 15 ml
- 1.5 ml Eppendorf DNA LoBind tubes
- 0.2 ml thin-walled PCR tubes
- Qubit™ Assay Tubes (Invitrogen, Q32856)
- Kit Qubit dsDNA HS (Invitrogen Q32851)
- Kit Qubit dsDNA BR (Invitrogen, Q32850)
Instrumental
- Dispositivo PromethION
- Pantalla protectora celdas de flujo PromethION
- Centrífuga y rotor adecuados para tubos Falcon de 15 ml
- Incubador o baño María ajustado a 37 °C y a 50 °C
- Asa bacteriológica o pinzas desechables
- Centrifugadora Eppendorf 5424 (o equivalente)
- Hula mixer (gentle rotator mixer)
- Magnetic separation rack, suitable for 1.5 ml Eppendorf tubes
- Microfuge
- Vortex mixer
- Thermal cycler
- Puntas de pipeta de orificio ancho
- P1000 pipette and tips
- P200 pipette and tips
- P100 pipette and tips
- P20 pipette and tips
- P10 pipette and tips
- P2 pipette and tips
- Ice bucket with ice
- Timer
Equipo opcional
- Sistema Agilent Femto Pulse (o equivalente)
- Qubit™ fluorometer (or equivalent for QC check)
La anterior lista de materiales, consumibles e instrumental corresponde al método de extracción de las secciones Preparación de muestras y Preparación de la biblioteca. Si cuenta con muestras ya extraídas, solo necesitará el material correspondiente a la sección Preparación de la biblioteca.
En este protocolo, se necesitan las siguientes entradas:
Requisitos de cantidad inicial por muestra en el método de extracción:
- 5 × 10⁶ de células por muestra
Cantidad de muestra necesaria por muestra en la preparación de la biblioteca:
- 2 μg de ADNg fragmentado en g-TUBE
Muestra inicial de ADN
Verificar la calidad de la muestra inicial de ADN
Es importante que la muestra de ADN cumpla con los requisitos de cantidad y calidad. Usar demasiado ADN, poco o de mala calidad (p. ej., que esté muy fragmentado, que contenga ARN o contaminantes químicos), tal vez afecte a la preparación de la biblioteca.
Siga las instrucciones del protocolo Input DNA/ RNA QC, sobre cómo realizar un control de calidad en la muestra de ADN.
Contaminantes químicos
Dependiendo de cómo se extraiga el ADN de la muestra sin procesar, ciertos contaminantes químicos pueden permanecer en el ADN purificado, lo cual tal vez afecte la eficacia de la preparación de la biblioteca y la calidad de la secuenciación. Encontrará más información sobre contaminantes en la página Contaminants de la comunidad Nanopore.
Reactivos de otros fabricantes
Hemos probado y recomendamos el uso de todos los reactivos de otros fabricantes usados en este protocolo. Oxford Nanopore Technologies no ha evaluado otras alternativas.
Recomendamos preparar estos reactivos siguiendo las instrucciones del fabricante.
Evaluación de la celda de flujo
Antes de empezar el experimento de secuenciación, recomendamos comprobar el número de poros disponibles, presentes en la celda de flujo. La evaluación deberá realizarse en los primeros tres meses desde su adquisición, si se trata de celdas de flujo MinION/GridION o PromethION. Oxford Nanopore Technologies sustituirá cualquier celda de flujo con un número de poros inferior al indicado en la tabla siguiente, siempre y cuando el resultado se notifique dentro de los dos días siguientes a la evaluación y se hayan seguido las instrucciones de almacenamiento. Para evaluar la celda de flujo, consultar las instrucciones en el documento Flow Cell Check.
| Celdas de flujo | Cantidad mínima de poros activos cubierta por la garantía |
|---|---|
| PromethION | 5 000 |
El adaptador incluido en este kit, Ligation Adapter (NA), no es intercambiable con otros adaptadores de secuenciación.
Contenido del kit Native Barcoding 24 V14 (SQK-NBD114.24)
Nota: estamos actualizando nuestros kits de códigos de barras sin amplificar (native barcoding), que ahora incluyen una mayor cantidad de Short Fragment Buffer (SFB). Si dispone de un kit en el formato antiguo y necesita una cantidad adicional de SFB, es posible adquirirla con el complemento SFB Expansion (EXP-SFB001).
Nuevo formato: mayor cantidad de Short Fragment Buffer (SFB)
| Nombre | Sigla | Color del tapón | Nº de viales | Volumen por vial (µl) |
|---|---|---|---|---|
| DNA Control Sample | DCS | Amarillo | 2 | 35 |
| Native Adapter | NA | Verde | 1 | 40 |
| Sequencing Buffer | SB | Rojo | 1 | 700 |
| Library Beads | LIB | Rosa | 1 | 600 |
| Library Solution | LIS | Tapón blanco, etiqueta rosa | 1 | 600 |
| Elution Buffer | EB | Negro | 2 | 500 |
| AMPure XP Beads | AXP | Tapón transparente, etiqueta verde claro | 1 | 6 000 |
| Long Fragment Buffer | LFB | Naranja | 1 | 1 800 |
| Short Fragment Buffer | SFB | Transparente | 1 | 13 000 |
| EDTA | EDTA | Azul | 1 | 700 |
| Flow Cell Flush | FCF | Tapón transparente, etiqueta azul celeste | 1 | 8 000 |
| Flow Cell Tether | FCT | Violeta | 1 | 200 |
| Native Barcode plate | NB01-24 | - | 2 placas, 3 juegos de códigos de barras por placa | 5 µl por pocillo |
Formato antiguo: menor cantidad de Short Fragment Buffer (SFB)
| Nombre | Sigla | Color del tapón | Nº de viales | Volumen por vial (µl) |
|---|---|---|---|---|
| DNA Control Sample | DCS | Amarillo | 2 | 35 |
| Native Adapter | NA | Verde | 1 | 40 |
| Sequencing Buffer | SB | Rojo | 1 | 700 |
| Library Beads | LIB | Rosa | 1 | 600 |
| Library Solution | LIS | Tapón blanco, etiqueta rosa | 1 | 600 |
| Elution Buffer | EB | Negro | 2 | 500 |
| AMPure XP Beads | AXP | Tapón transparente, etiqueta verde claro | 1 | 6 000 |
| Long Fragment Buffer | LFB | Naranja | 1 | 1 800 |
| Short Fragment Buffer | SFB | Transparente | 1 | 1 800 |
| EDTA | EDTA | Azul | 1 | 700 |
| Flow Cell Flush | FCF | Tapón transparente, etiqueta azul celeste | 1 | 8 000 |
| Flow Cell Tether | FCT | Violeta | 1 | 200 |
| Native Barcode plate | NB01-24 | - | 2 placas, 3 juegos de códigos de barras por placa | 5 µl por pocillo |
Nota: este producto contiene reactivo AMPure XP, fabricado por Beckman Coulter, Inc., y puede almacenarse a −20 °C junto con el kit sin comprometer su estabilidad.
Nota: la muestra de control de ADN (DCS) es un amplicón estándar de 3,6 kb que corresponde al extremo 3’ del genoma Lambda.
3. Archivo BED
Descargar el archivo BED del catálogo Muestreo adaptativo.
El catálogo de muestreo adaptativo facilita que tanto el equipo de Oxford Nanopore como los miembros de la comunidad compartan archivos BED con regiones genómicas seleccionadas, utilizadas en experimentos de muestreo adaptativo. Los archivos BED junto al genoma de referencia se cargan en MinKNOW.
En los experimentos de RRMS con genoma humano, descargar el archivo Secuenciación de metilación de representación reducida humana (RRMS)
En experimentos de RRMS con genoma de ratones, descargar el archivo Secuenciación de metilación de representación reducida en ratones (RRMS).
(Opcional): en experimentos con genomas de otros vertebrados, utilizar un archivo BED a medida para el organismo deseado.
4. Extracción de ADN
Material
- 5 × 10⁶ de células
Consumibles
- Kit Puregene Cell (QIAGEN, 158043)
- Etanol al 70 % recién preparado en agua sin nucleasas
- Tampón TE (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) (Fisher Scientific, 10224683)
- PBS 1× (solución salina tamponada con fosfato)
- Isopropanol
- Kit Qubit dsDNA HS (ThermoFisher, Q32851)
- Qubit™ Assay Tubes (Invitrogen, Q32856)
- Tubos Falcon de 15 ml
- 1.5 ml Eppendorf DNA LoBind tubes
Instrumental
- Centrífuga y rotor adecuados para tubos Falcon de 15 ml
- Incubador o baño María ajustado a 37 °C y a 50 °C
- Vortex mixer
- Asa bacteriológica o pinzas desechables
- Puntas de pipeta de orificio ancho
- P1000 pipette and tips
- P200 pipette and tips
- P100 pipette and tips
- P20 pipette and tips
- Qubit™ fluorometer (or equivalent for QC check)
Extracción a partir de estirpes celulares cultivadas:
Extraer el ADN de la(s) muestra(s) siguiendo uno de los protocolos de extracción recomendados.
Como parte de la evaluación comparativa de este método, el equipo de Oxford Nanopore extrajo ADN de ~5 millones de células utilizando el protocolo: ADN de estirpes celulares humanas – kit Puregene Cell de QIAGEN. A continuación, describimos los pasos de este método.
Nota: este método también admite ADN de estirpes celulares de ratón.
También ofrecemos varios protocolos de extracción de muestras de mamíferos, que es posible utilizar con otros tipos de muestras.
Recoger y sedimentar 5 × 10⁶ de células mediante centrifugación a 300 × g (RCF) durante 3 minutos. Si queda líquido junto al sedimento, centrifugar las células de nuevo y aspirar el resto del sobrenadante.
Añadir 200 μl de PBS 1× al ADN sedimentado y centrifugar a 300 x g durante 3 minutos. Aspirar y desechar el sobrenadante.
Añadir 2 ml de solución de lisis celular al ADN sedimentado. Con una punta de pipeta de punta ancha, resuspender las células y transferirlas a un tubo Falcon de 15 ml. Si quedan grumos de células, invertir el tubo con suavidad.
Incubar la muestra a 37 °C durante 30 minutos.
Añadir 700 μl de solución de precipitante de proteínas a las células lisadas y mezclar mediante vórtex durante 3 ciclos de 5 segundos.
Centrifugar la muestra a 2 000 x g durante 5 minutos.
Transferir el sobrenadante a un tubo nuevo y añadir 2,5 ml de isopropanol a temperatura ambiente. Desechar el sedimento.
Mezclar el contenido invirtiendo el tubo con suavidad 50 veces.
Enrollar el ADN con un asa bacteriológica.
Sumergir el ADN enrollado en un tubo Eppendorf que contenga etanol frío al 70 %.
Retirar el asa bacteriológica con el ADN enrollado del tubo de etanol y dejar secar al aire durante unos segundos.
Sumergir el ADN en un tubo Eppendorf DNA LoBind de 1,5 ml que contenga 250 μl TE (1mM EDTA, pH 8,0) y dejar que el ADN se desprenda suavemente del asa.
Incubar el tubo durante 2 horas a 50 °C y mezclar de vez en cuando el contenido del tubo (200 μl) con una pipeta de punta ancha.
Nota: el sedimento de ADN tal vez tarde un poco en solubilizarse. Compruebe que la solución esté bien mezclada antes de cuantificarla.
Opcional: otra posibilidad es realizar la incubación a temperatura ambiente durante la noche.
Cuantificar 1 μl de cada muestra eluida utilizando un fluorímetro Qubit.
Tomar 2 μg del ADNg extraído por cada muestra y proceder a la fragmentación del ADN.
5. Fragmentación de ADN
Material
- 2 µg de ADNg extraído (en el paso anterior)
Consumibles
- Tubo g-TUBE™ (Covaris, 520079)
- Tampón TE (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) (Fisher Scientific, 10224683)
- Kit Qubit dsDNA BR (Invitrogen, Q32850)
- Qubit™ Assay Tubes (Invitrogen, Q32856)
- 1.5 ml Eppendorf DNA LoBind tubes
Instrumental
- Centrifugadora Eppendorf 5424 (o equivalente)
- P1000 pipette and tips
- P200 pipette and tips
- P100 pipette and tips
- P20 pipette and tips
- P2 pipette and tips
- Qubit™ fluorometer (or equivalent for QC check)
Equipo opcional
- Sistema Agilent Femto Pulse (o equivalente)
Fragmentación de ADN extraído con un tubo g-TUBE de Covaris:
La fragmentación mecánica del ADN genómico se lleva a cabo con un tubo g-TUBE (Covaris), que permite cizallar el ADN y obtener fragmentos de aproximadamente 6 kb, como paso previo al protocolo de preparación de la biblioteca.
Preparar al ADN en tampón TE:
- Comprobar que tiene 2 μg de ADNg extraído de la muestra y transferirlo a un tubo Eppendorf de 1,5 ml.
- Ajustar el volumen a un total de 50 μl con tampón TE.
- Mezclar minuciosamente aspirando y expulsando con la pipeta.
- Centrifugar brevemente en la microcentrífuga.
Cargar los 50 μl de la muestra en la parte superior del tubo g-TUBE. Enroscar la tapa con firmeza y centrifugar a 11 000 rpm (~11 300 RCF) durante 30 segundos.
A continuación, centrifugar el tubo de nuevo a 11 000 rpm (~11 300 RCF) durante 10 segundos, hasta que todo el ADNg pase por el estrechamiento.
Comprobar que la muestra haya pasado por completo de la cámara superior a la cámara inferior del tubo g-TUBE.
Invertir el tubo g-TUBE y centrifugar de nuevo a la misma velocidad y duración que en el paso anterior: 11 000 rpm (~11 300 RCF) durante 30 segundos.
Repetir la centrifugación a 11 000 rpm (~11 300 RCF) durante 10 segundos, hasta que todo el ADNg pase por el estrechamiento.
Desenroscar el cuerpo del tubo. La tapa roscada, que contiene la muestra, quedará expuesta. Tomar la muestra y transferirla a un tubo Eppendorf nuevo de 1,5 ml.
Cuantificar 1 μl del ADNg fragmentado con el kit de ensayo Qubit dsDNA Broad Range.
Se espera que la concentración de la muestra, tras el cizallamiento del g-TUBE, sea de ~40 ng/µl.
El ADNg fragmentado también debe evaluarse con el sistema de electroforesis capilar Femto-Pulse (Agilent), que analiza el tamaño y la calidad del ADN.

Distribución representativa de fragmentos de ADN tras la fragmentación con g-TUBE, analizada con el ensayo Femto-Pulse de Agilent (hasta 165 kb). Obsérvese un único pico prominente de aproximadamente 6 kb.
Tomar 48 μl de ADNg fragmentado (~2 µg por muestra) y continuar con la preparación de la biblioteca.
6. Reparación del ADN y preparación de los extremos
Material
- 2 µg de ADNg fragmentado en 48 µl por muestra
- AMPure XP Beads (AXP)
Consumibles
- NEBNext FFPE DNA Repair Mix (NEB M6630)
- Módulo NEBNext® Ultra™ II End Repair/dA-Tailing (NEB, E7546)
- Kit Qubit dsDNA HS (Invitrogen Q32851)
- Nuclease-free water (e.g. Thermo Scientific, AM9937)
- Etanol al 80 % recién preparado con agua sin nucleasas
- Qubit™ Assay Tubes (Invitrogen, Q32856)
- 0.2 ml thin-walled PCR tubes
- 1.5 ml Eppendorf DNA LoBind tubes
Instrumental
- P1000 pipette and tips
- P100 pipette and tips
- P10 pipette and tips
- Microfuge
- Thermal cycler
- Hula mixer (gentle rotator mixer)
- Gradilla magnética
- Ice bucket with ice
Equipo opcional
- Qubit™ fluorometer (or equivalent for QC check)
Preparar los reactivos NEBNext FFPE DNA Repair Mix y NEBNext Ultra II End Repair / dA-tailing Module siguiendo las instrucciones del fabricante y poner en hielo.
A fin de obtener un rendimiento óptimo, NEB recomienda lo siguiente:
Descongelar todos los reactivos en hielo.
Golpear suavemente los tubos de los reactivos con el índice o invertirlos hasta que estén bien mezclados. No mezclar en vórtex las mezclas FFPE DNA Repair Mix, ni Ultra II End Prep Enzyme Mix.
Centrifugar brevemente los tubos antes de abrirlos.
Los tampones Ultra II End Prep Reaction Buffer y FFPE DNA Repair Buffer tal vez tengan un poco de precipitado. Dejar que la mezcla alcance la temperatura ambiente y mezclar con la pipeta varias veces hasta homogeneizarla; agitar mediante vórtex durante 30 segundos para solubilizarla.
Nota es importante mezclar bien los tampones mediante vórtex.El tampón FFPE DNA Repair Buffer tal vez tenga un matiz amarillo; no importa si está así; se puede utilizar.
Preparar las muestras de ADN en agua sin nucleasas:
Para cada muestra, se necesitan ~2 μg de ADN extraído y fragmentado en tubos independientes Eppendorf DNA LoBind de 1,5 ml.
Si es necesario, ajustar el volumen a un total de 48 μl con agua sin nucleasas.
Mezclar minuciosamente con la pipeta o golpear el tubo suavemente con el índice.
Centrifugar brevemente en la microcentrífuga.
En cada uno de los tubos Eppendorf de 1,5 ml, que contienen las muestras de ADN fragmentado, mezclar lo siguiente:
| Reactivo | Volumen |
|---|---|
| ~2 µg de ADN fragmentado del paso anterior | 48 µl |
| Ultra II End-prep Reaction Buffer | 3,5 µl |
| Ultra II End-prep Enzyme Mix | 3 µl |
| NEBNext FFPE DNA Repair Buffer | 3,5 µl |
| NEBNext FFPE DNA Repair Mix | 2 µl |
| Total | 60 µl |
Mezclar minuciosamente la reacción con la pipeta y centrifugar brevemente.
En un termociclador con tapa térmica, incubar la reacción a 20 °C durante 5 minutos, a 65 °C durante 5 minutos y mantener a 4 °C.
Sacar la reacción del termociclador y colocar el tubo en hielo.
Resuspender las microesferas AMPure XP (AXP) mediante vórtex.
Añadir un volumen (60 µl) de microesferas AMPure XP (AXP) resuspendidas a la reacción de preparación de extremos y mezclar dando suaves golpes en el tubo.
Incubar en el hula mixer (o mezclador rotatorio) durante 5 minutos a temperatura ambiente.
Preparar 3 ml de etanol al 80 % con agua sin nucleasas.
Centrifugar brevemente la muestra y sedimentar en una gradilla magnética hasta que el sobrenadante se vuelva claro e incoloro. Sin mover el tubo, retirar el sobrenadante con una pipeta.
Con el tubo en la gradilla, lavar las microesferas con 200 µl de etanol al 80 %, sin desplazar el sedimento. Retirar el etanol con una pipeta y desechar.
Si se altera el sedimento, esperar que las microesferas sedimenten de nuevo antes de retirar el etanol.
Repetir el paso anterior.
Centrifugar brevemente y colocar el tubo de nuevo en la gradilla magnética. Retirar con una pipeta cualquier residuo de etanol. Dejar secar el sedimento durante 30 segundos aproximadamente, sin dejar que se agriete.
Quitar el tubo de la gradilla magnética y resuspender el sedimento en 20 µl de agua sin nucleasas (repetir el proceso con cada muestra). Incubar durante 2 minutos a temperatura ambiente.
Sedimentar las microesferas en una gradilla magnética, durante al menos 1 minuto, hasta que el eluido se vuelva claro e incoloro.
Extraer 20 µl del eluido de cada muestra y guardarlo en un tubo Eppendorf DNA Lobind de 1,5 ml.
Nota: procesar las muestras por separado. En esta fase, aún no tienen códigos de barras.
Cuantificar cada muestra eluida con un fluorímetro Qubit.
Nota: recomendamos realizar tres lecturas de Qubit por cada una de las muestras, a fin de cuantificarlas con mayor precisión, lo que será esencial para la posterior normalización de cada muestra antes de añadir los códigos.
Tras la preparación de extremos se recuperarán entre 1000 y 1600 ng por muestra.
A partir de los resultados de cuantificación, normalizar las muestras con la muestra de menor rendimiento.
- Extraer 15 μl de la muestra de menor rendimiento y poner en un tubo nuevo de PCR de pared fina (0,2 ml).
- Extraer una masa equivalente de cada una de las otras muestras y poner en tubos nuevos, separados, de PCR de pared fina (0,2 ml).
- Ajustar el volumen de cada una de las muestras a un total de 15 μl con agua sin nucleasas.
Usar 15 μl de cantidad equimolar de las muestras de ADN reparado y llevar a cabo la ligación de códigos de barras sin realizar amplificación. En este paso, también se puede guardar la muestra a 4 °C hasta el día siguiente.
7. Ligación de los códigos de barras sin amplificación
Material
- ADN reparado del paso anterior en un volumen de 15 µl
- Códigos de barras sin amplificación - Native barcodes (NB01-24)
- AMPure XP Beads (AXP)
- EDTA (ácido etilendiaminotetraacético)
- Short Fragment Buffer (SFB)
Consumibles
- NEB Blunt/TA Ligase Master Mix (NEB, M0367)
- Etanol al 80 % recién preparado con agua sin nucleasas
- Nuclease-free water (e.g. Thermo Scientific, AM9937)
- 1.5 ml Eppendorf DNA LoBind tubes
- Placa de PCR Eppendorf twin.tec® LoBind de 96 pocillos, con semifaldón y sellado térmico (Eppendorf™, 0030129504)
- o tubos de PCR de pared fina de 0,2 ml
- Qubit™ Assay Tubes (Invitrogen, Q32856)
- Kit Qubit dsDNA HS (ThermoFisher, Q32851)
Instrumental
- Gradilla magnética
- Vortex mixer
- Hula mixer (gentle rotator mixer)
- Microfuge
- Thermal cycler
- Ice bucket with ice
- Multichannel pipette and tips
- P1000 pipette and tips
- P200 pipette and tips
- P100 pipette and tips
- P20 pipette and tips
- P10 pipette and tips
- P2 pipette and tips
- Qubit™ fluorometer (or equivalent for QC check)
Preparar los reactivos NEB Blunt/TA Ligase Master Mix siguiendo las instrucciones del fabricante y colocar en hielo:
Descongelar los reactivos a temperatura ambiente.
Centrifugar brevemente los viales durante 5 segundos.
Mezclar los reactivos con la pipeta al menos 10 veces hasta que estén completamente homogéneos.
Descongelar el EDTA a temperatura ambiente y mezclar en vórtex. Centrifugar brevemente y colocar en hielo.
Descongelar los códigos de barras sin amplificar (NB01-24) a temperatura ambiente. Centrifugar brevemente y con la pipeta, mezclar uno a uno los códigos necesarios según el número de muestras. Colocar en hielo.
Los pocillos de la placa con códigos de barras son de un solo uso. Sellar los pocillos antes de utilizarlos y no reutilizarlos una vez abiertos o perforados.
Seleccionar un código de barras distinto para cada muestra que se vaya a procesar en la misma celda de flujo. Las muestras deben llevar códigos de barras y estar agrupadas en el mismo experimento.
Nota: utilizar solo un código de barras por muestra.
En los tubos de PCR de 0,2 ml que contienen las muestras normalizadas, añadir los siguientes reactivos a cada muestra, en este orden:
| Reactivo | Volumen |
|---|---|
| ADN reparado | 15 µl |
| Códigos de barras sin amplificar (NB01-24) | 5 µl |
| Blunt/TA Ligase Master Mix | 20 µl |
| Total | 40 µl |
Mezclar bien la reacción con la pipeta y centrifugar brevemente.
Incubar durante 20 minutos a temperatura ambiente.
Añadir 4 µl de EDTA (tapón azul) a cada pocillo, mezclar bien con la pipeta y centrifugar brevemente.
En este paso se añade EDTA, que detiene la reacción.
Agrupar las muestras con códigos de barras en un tubo Eppendorf DNA LoBind de 1,5 ml.
| . | 4 muestras |
|---|---|
| Volumen total en preparaciones con EDTA (tapón azul) | 176 µl |
Recomendamos comprobar que la base de los tubos/placa tenga el mismo volumen antes y después de agruparlos y así verificar que se haya extraído todo el líquido.
Resuspender las microesferas AMPure XP (AXP) mediante vórtex.
Añadir 0,65× de microesferas AmPure XP (AXP) a la reacción y mezclar con la pipeta.
| . | 4 muestras |
|---|---|
| Volumen de AXP en preparaciones con EDTA (tapón azul) | 115 µl |
Incubar en el hula mixer (o mezclador rotatorio) durante 10 minutos a temperatura ambiente.
En el siguiente paso de purificación se utiliza Short Fragment Buffer (SFB) en lugar de etanol al 80 % para lavar las microesferas. El uso de etanol sería perjudicial para la reacción de secuenciación.
Centrifugar brevemente la muestra y sedimentar en una gradilla magnética durante 5 minutos. Esperar hasta que el eluido se vuelva claro e incoloro y retirar el sobrenadante con una pipeta.
Lavar las microesferas con 500 μl de Short Fragment Buffer (SFB). Golpear el tubo suavemente con el dedo, centrifugar brevemente, colocar de nuevo en la gradilla magnética y dejar que las microesferas sedimenten. Retirar el sobrenadante con una pipeta y desechar.
Repetir el paso anterior.
Centrifugar brevemente y colocar el tubo de nuevo en la gradilla magnética. Retirar con la pipeta cualquier resto de Short Fragment Buffer (SFB). Dejar secar el sedimento durante 30 segundos aproximadamente, sin dejar que se agriete.
Quitar el tubo de la gradilla magnética y resuspender el sedimento en 32 µl de agua sin nucleasas dando suaves golpes al tubo con el dedo.
Incubar durante 15 minutos a 37 °C. Cada 2 minutos, agitar la muestra dando suaves golpes al tubo con el dedo durante 10 segundos, a fin de favorecer la elución de ADN.
Sedimentar las microesferas en una gradilla magnética hasta que el eluido se vuelva claro e incoloro.
Extraer y guardar 32 µl de eluido en un tubo nuevo Eppendorf DNA Lobind de 1,5 ml.
Cuantificar 1 μl de muestra eluida con un fluorímetro Qubit.
Nota: tras la ligación de los códigos de barras, por lo general, se recuperan entre 2200 y 3200 ng.
Tomar la biblioteca de ADN obtenida y proceder a la ligación de los adaptadores y purificación. No obstante, es posible conservar la muestra a 4 °C durante la noche.
8. Ligación de los adaptadores y purificación
Material
- Long Fragment Buffer (LFB)
- Elution Buffer (EB)
- Native Adapter (NA)
- AMPure XP Beads (AXP)
Consumibles
- NEBNext® Quick Ligation Module (NEB, E6056)
- 1.5 ml Eppendorf DNA LoBind tubes
- Qubit™ Assay Tubes (Invitrogen, Q32856)
- Kit Qubit dsDNA HS (ThermoFisher, Q32851)
Instrumental
- Microfuge
- Gradilla magnética
- Vortex mixer
- Hula mixer (gentle rotator mixer)
- Thermal cycler
- P1000 pipette and tips
- P200 pipette and tips
- P100 pipette and tips
- P20 pipette and tips
- P10 pipette and tips
- Ice bucket with ice
- Qubit™ fluorometer (or equivalent for QC check)
El Native Adapter (NA) utilizado en este kit y protocolo no es intercambiable con otros adaptadores de secuenciación.
Verificar la celda de flujo
Antes de empezar a preparar la biblioteca, recomendamos verificar la celda de flujo para comprobar que tiene poros suficientes para realizar un buen experimento.
Las instrucciones de comprobación de la celda de flujo están disponibles en el protocolo de MinKNOW.
Preparar el reactivo NEBNext Quick Ligation Reaction Module de acuerdo con las instrucciones del fabricante y colocar en hielo:
Descongelar los reactivos a temperatura ambiente.
Centrifugar los tubos de reactivos durante 5 segundos.
Mezclar los reactivos con la pipeta al menos 10 veces hasta que estén completamente homogéneos.
Nota: no mezclar en vórtex la ligasa T4 DNA.
El reactivo NEBNext Quick Ligation Reaction Buffer (5x) quizás tenga un poco de precipitado. Dejar que la mezcla alcance la temperatura ambiente y mezclar con la pipeta varias veces hasta homogeneizarlo; agitar en vórtex durante varios segundos hasta que el reactivo esté completamente mezclado.
No mezclar en vórtex la ligasa T4 DNA.
Centrifugar brevemente los viales Ligation Adapter (NA) y Quick T4 Ligase, mezclar con la pipeta y poner en hielo.
Descongelar el tampón Elution Buffer (EB) a temperatura ambiente y mezclar en vórtex. Centrifugar brevemente y poner en hielo.
Descongelar el tampón Long Fragment Buffer LFB a temperatura ambiente y mezclar en vórtex. Centrifugar brevemente y poner en hielo.
En un tubo de 1,5 ml Eppendorf DNA LoBind, mezclar en el siguiente orden:
Entre cada adición, mezclar con la pipeta de 10 a 20 veces.
| Reactivo | Volumen |
|---|---|
| Agrupación de muestras | 30 µl |
| Native Adapter (NA) | 5 µl |
| NEBNext Quick Ligation Reaction Buffer (5×) | 10 µl |
| Quick T4 DNA Ligase | 5 µl |
| Total | 50 µl |
Mezclar bien la reacción con la pipeta y centrifugar brevemente.
Incubar la reacción durante 20 minutos a temperatura ambiente.
En el siguiente paso de purificación se utiliza Short Fragment Buffer (LFB) en lugar de etanol al 80 % para lavar las microesferas. El uso de etanol sería perjudicial para la reacción de secuenciación.
Resuspender las microesferas AMPure XP (AXP) mediante vórtex.
Añadir 25 μl (0,5 x) de microesferas AmPure XP (AXP) resuspendidas a la mezcla de reacciones y mezclar con la pipeta.
Incubar en el hula mixer (o mezclador rotatorio) durante 10 minutos a temperatura ambiente.
Centrifugar brevemente la muestra y sedimentar en una gradilla magnética. Sin mover el tubo, retirar el sobrenadante con una pipeta.
Lavar las microesferas con 250 μl de Long Fragment Buffer (LFB). Golpear el tubo suavemente con el dedo, centrifugar brevemente, colocar de nuevo en la gradilla magnética y dejar que las microesferas sedimenten durante al menos 5 minutos. Retirar el sobrenadante con una pipeta y desechar.
Nota: retirar el sobrenadante con cuidado; la viscosidad del tampón tal vez contribuya a que se pierdan microesferas del sedimento.
Repetir el paso anterior.
Centrifugar brevemente y colocar el tubo de nuevo en la gradilla magnética. Retirar con una pipeta cualquier residuo de sobrenadante. Dejar secar el sedimento durante 30 s aproximadamente, sin dejar que se agriete.
Quitar el tubo de la gradilla magnética y resuspender el sedimento en 97 µl de Elution Buffer (EB).
Centrifugar brevemente e incubar durante 20 minutos a 37 °C. Cada 2 minutos, dar suaves golpes al tubo con el dedo durante 10 segundos y así favorecer la elución de ADN.
Sedimentar las microesferas en una gradilla magnética, durante al menos 1 minuto, hasta que el eluido se vuelva claro e incoloro.
Extraer y conservar 97 μl del eluido que contiene la biblioteca de ADN en un tubo nuevo de 1,5 ml Eppendorf DNA LoBind.
Deshechar las microesferas sedimentadas.
Cuantificar 1 μl de muestra eluida utilizando un fluorímetro Qubit.
Nota: en un volumen de 96 μl, por lo general, se recuperan entre 1000 y 1200 ng tras la ligación y purificación.
La biblioteca preparada se usará para cargar la celda de flujo. Conservar la biblioteca en hielo o a 4 °C hasta el momento de cargar.
Recomendaciones de almacenamiento de la biblioteca
Recomendamos guardar las bibliotecas en tubos Eppendorf DNA LoBind a 4 °C, durante periodos de tiempo cortos o en caso de uso repetido, por ejemplo, para recargar celdas de flujo entre lavados. Para uso individual y conservar a largo plazo durante periodos de más de 3 meses, recomendamos guardar las bibliotecas a –80 °C en tubos Eppendorf DNA LoBind.
9. Acondicionamiento y carga de la celda de flujo PromethION.
Material
- Sequencing Buffer (SB)
- Library Beads (LIB)
- Flow Cell Tether (FCT)
- Flow Cell Flush (FCF)
Consumibles
- Celdas de flujo PromethION
- 1.5 ml Eppendorf DNA LoBind tubes
Instrumental
- Dispositivo PromethION 24/48 o PromethION 2 Solo
- Dispositivo PromethION
- Pantalla protectora celdas de flujo PromethION
- P1000 pipette and tips
- P200 pipette and tips
- P20 pipette and tips
Este kit es compatible solo con las celdas de flujo R10.4.1 (FLO-PRO114M).
Al sacar las celdas de flujo de la nevera, esperar 20 minutos antes de insertarlas en el dispositivo, y así darles tiempo a que alcancen la temperatura ambiente. En ambientes húmedos puede formarse condensación en la celda de flujo. Inspeccione las clavijas doradas del conector, situadas en la parte superior e inferior de la celda de flujo, en busca de condensación y si la hubiera, límpiela con una toallita sin pelusa. Compruebe que haya una almohadilla térmica negra enganchada en la parte posterior de la celda de flujo.
Descongelar los viales Sequencing Buffer (SB), Library Beads (LIB), Flow Cell Tether (FCT) y Flow Cell Flush (FCF) a temperatura ambiente y mezclar en vórtex. Centrifugar brevemente y poner en hielo.
Preparar la mezcla de acondicionamiento; para ello, combinar Flow Cell Tether (FCT) y Flow Cell Flush (FCF) como se indica a continuación. Agitar en vórtex a temperatura ambiente.
En un tubo proporcionado al número de celdas de flujo que se vayan a utilizar, combinar los siguientes reactivos:
| Reactivo | Volumen por celda de flujo |
|---|---|
| Flow Cell Flush (FCF) | 1 170 µl |
| Flow Cell Tether (FCT) | 30 µl |
| Total volume | 1 200 µl |
En el PromethION 2 Solo, cargar la(s) celda(s) de flujo de la siguiente manera:
Colocar la celda de flujo sobre la placa metálica.
Deslizarla hacia el puerto de acople hasta que no se vean las clavijas doradas o la placa verde.

En el PromethION 24/48 cargar la(s) celda(s) de flujo en el/los puerto(s) de acople:
- Alinear la celda de flujo con el conector (horizontal y verticalmente) e insertarla suavemente en la posición.
- Presionar la celda de flujo con firmeza y comprobar que el cierre encaje y haga clic.


Insertar la celda de flujo en el ángulo equivocado puede dañar las clavijas del PromethION y afectar los resultados de secuenciación. Si observa que las clavijas de las posiciones del PromethION están dañadas, contacte con el servicio de asistencia en support@nanoporetech.com.

Antes de cargar la biblioteca, verificar la celda de flujo y comprobar el número de poros que tiene.
Este paso puede omitirse si la celda de flujo se ha comprobado con anterioridad.
Para más información, consultar las instrucciones de comprobación de la celda de flujo en el protocolo de MinKNOW.
Deslizar el puerto de entrada en el sentido de las agujas del reloj.

Tenga cuidado al extraer el tampón de la celda de flujo. No retire más de 20-30 μl y asegúrese de que cubra la matriz de poros en todo momento. La introducción de burbujas de aire en la matriz puede dañar los poros de manera irreversible.
Tras abrir el puerto de entrada, retirar una pequeña cantidad de tampón para quitar las burbujas de aire:
- Ajustar una pipeta P1000 a 200 μl.
- Introducir la punta de la pipeta en el puerto de entrada.
- Girar la rueda hasta que el indicador de volumen marque 220-230 μl o hasta que se pueda ver una pequeña cantidad entrar en la punta de la pipeta.

Cargar 500 μl de mezcla de acondicionamiento por el puerto de entrada, evitando introducir burbujas de aire. Esperar cinco minutos. Durante este tiempo, preparar la biblioteca para cargar siguiendo los siguientes pasos del protocolo.

Mezclar minuciosamente con la pipeta el contenido del vial Library Beads (LIB).
El vial Library Beads (LIB) contiene microesferas en suspensión. Las microesferas sedimentan muy rápido; por eso es crucial mezclarlas justo antes de su uso.
Recomendamos utilizar Library Beads (LIB) en la mayoría de experimentos de secuenciación. No obstante, el reactivo Library Solution (LIS) está disponible en caso de que se utilicen bibliotecas más viscosas.
En un tubo Eppendorf DNA LoBind de 1,5 ml preparar la biblioteca de la siguiente manera:
| Reactivo | Volumen por celda de flujo |
|---|---|
| Sequencing Buffer (SB) | 100 µl |
| Library Beads (LIB) bien mezclado antes de su uso | 68 µl |
| Biblioteca de ADN | 32 µl |
| Total | 200 µl |
Nota: la biblioteca preparada se utiliza para cargar la celda de flujo. Conservar la biblioteca en hielo o a 4 °C hasta el momento de cargar.
Cargar 500 μl de mezcla de acondicionamiento por el puerto de entrada y así finalizar el acondicionamiento de la celda de flujo.

Mezclar la biblioteca suavemente con la pipeta justo antes de cargarla.
Cargar 200 μl de biblioteca por el puerto de entrada con una pipeta p1000.

Cerrar la válvula. De este modo se sellará el puerto de entrada.

A fin de lograr un rendimiento de secuenciación óptimo, colocar la pantalla protectora sobre la celda de flujo tan pronto como se haya cargado la biblioteca.
Recomendamos no retirar la pantalla protectora de la celda de flujo mientras la biblioteca esté cargada, incluso durante los lavados o recargas.
Si se ha retirado la pantalla protectora de la celda de flujo, se instala de la siguiente manera:
- Alinear la abertura de la pantalla protectora con la tapa del puerto de entrada. El borde delantero de la pantalla protectora debe quedar por encima del identificador de la celda de flujo.
- Presionar la pantalla protectora con firmeza alrededor de la tapa del puerto de entrada. Cuando el clip del puerto de entrada encaje bajo la tapa del mismo, se producirá un clic.
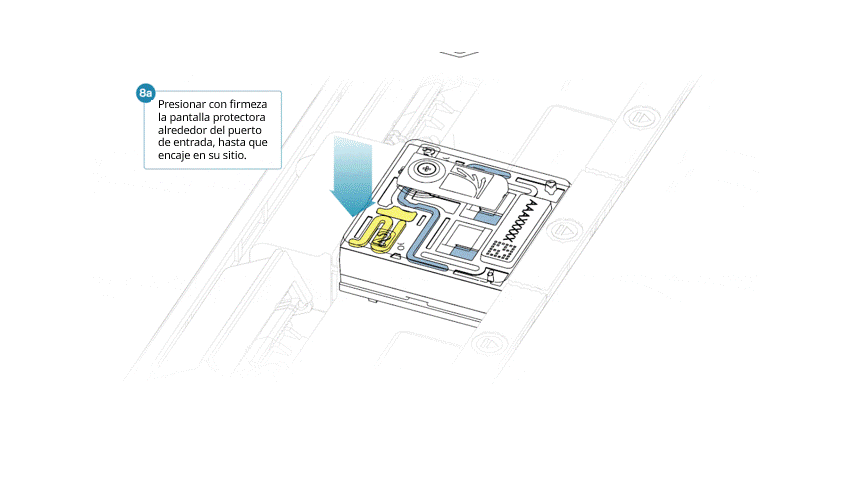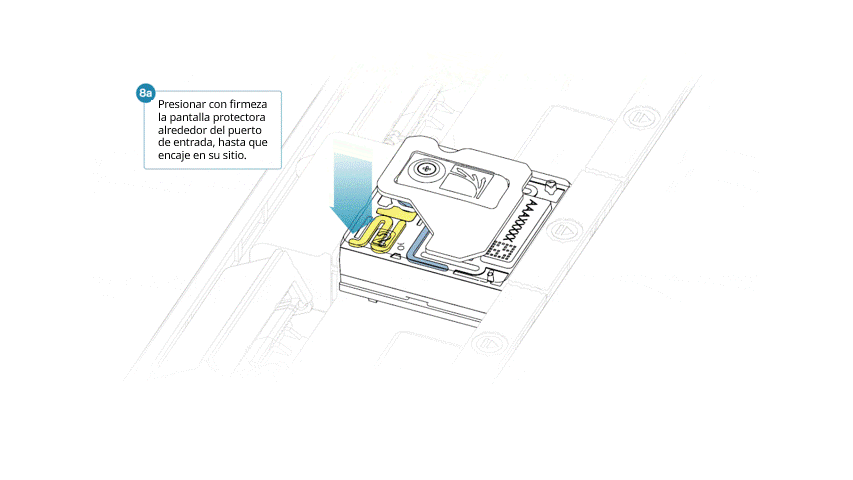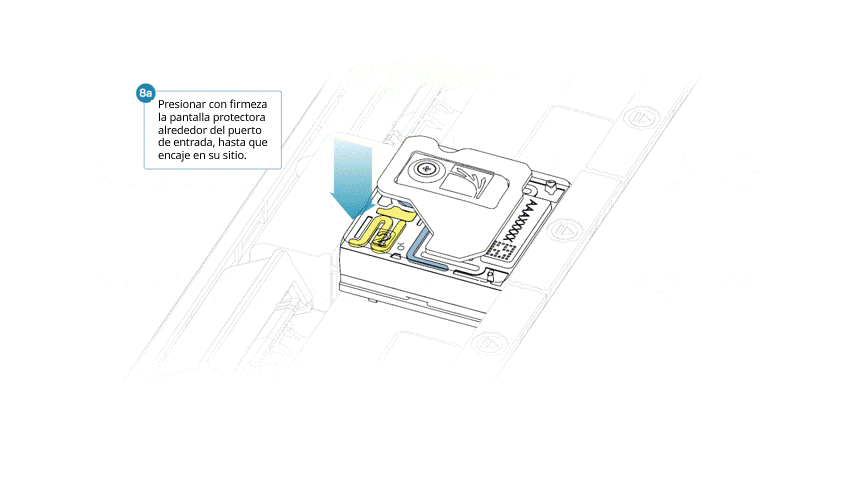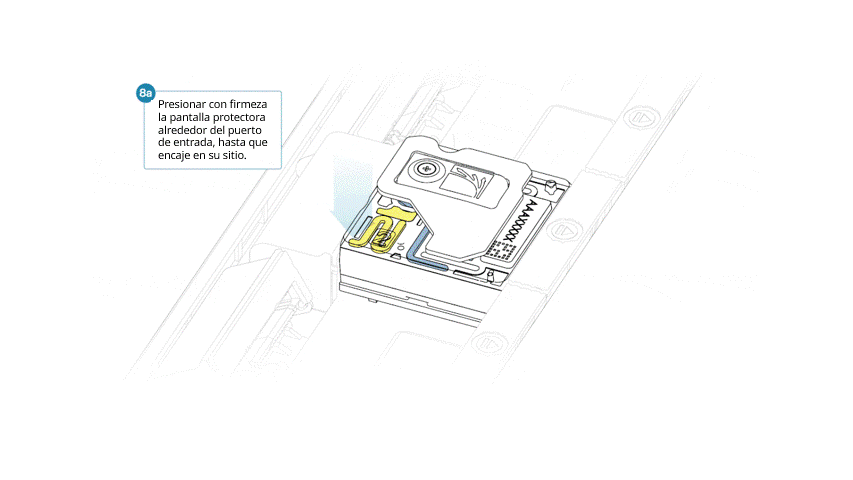

Cerrar la tapa del PromethION cuando esté listo para empezar un experimento de secuenciación en MinKNOW.
Esperar al menos 10 minutos tras cargar las celdas de flujo en el PromethION antes de iniciar cualquier experimento ayudará a aumentar el rendimiento de la secuenciación.
Las instrucciones para configurar los experimentos de secuenciación están en la sección Adquisición de datos e identificación de bases de este protocolo.
Recordatorio en este protocolo, recomendamos lavar y volver a cargar la celda de flujo con biblioteca nueva para mantener elevada la adquisición de datos tras aproximadamente 24 h de secuenciación.
Siga las instrucciones de la sección «Lavado y recarga de celdas de flujo PromethION» de este protocolo.
10. Lavado y recarga de celdas de flujo PromethION
Material
- Biblioteca de ADN con adaptadores (sección anterior)
- Kit Flow Cell Wash (EXP-WSH004)
- Sequencing Auxiliary Vials V14 (EXP-AUX003)
Consumibles
- 1.5 ml Eppendorf DNA LoBind tubes
Instrumental
- P1000 pipette and tips
- P20 pipette and tips
- Ice bucket with ice
- Vortex mixer
Recomendamos lavar y volver a cargar la celda de flujo después de aproximadamente 24 h de secuenciación.
En este método, la celda de flujo se lava tras 24 h de secuenciación para restablecer los poros y garantizar una adquisición de datos eficaz. Tras 24 h de secuenciación adicionales, se lava la celda de flujo y se vuelve a cargar por segunda vez. Por este motivo se creó suficiente biblioteca para cargar 3 celdas de flujo, en la fase de ligación del adaptador.
- Este método de lavado tiene por objeto eliminar la mayor parte de la biblioteca inicial y desbloquear los poros como preparación para cargar la biblioteca siguiente.
- La adquisición de datos en MinKNOW debe interrumpirse durante el lavado y la carga de la biblioteca.
- Una vez lavada la celda de flujo, es posible cargar la siguiente biblioteca.
Navegar hasta el gráfico de actividad o el de resultados de exploración para ver la disponibilidad de poros.
A continuación, se presentan datos de ejemplo sobre el estado de los poros observados en una celda de flujo antes y después de realizar los pasos de lavado. Además, es posible observar un ejemplo de los datos de secuenciación acumulados, incluyendo los pasos de lavado y recarga. Los asteriscos rojos indican los lavados y las recargas de la celda de flujo.

Imagen 1. Estado del canal durante un experimento de secuenciación de 96 horas. Los lavados de la celda de flujo están incluidos en el método para restablecer los poros bloqueados y permitir una adquisición continua de datos. Los asteriscos rojos indican que se ha realizado un purgado.

Imagen 2. Estado del canal durante un experimento de secuenciación de 96 horas. Los asteriscos rojos indican que se ha realizado un purgado.
Vídeo: lavado y recarga de celdas de flujo PromethION
Este vídeo muestra cómo lavar una celda de flujo tras un experimento de secuenciación y cómo cargar una nueva biblioteca.
Recomendamos mantener la pantalla protectora en la celda de flujo durante el lavado, si se va a cargar de inmediato una segunda biblioteca.
Si se va a lavar y guardar la celda de flujo, es posible retirar la pantalla protectora.
Colocar el tubo Wash Mix (WMX) en hielo. No mezclar en vórtex.
Descongelar un vial de Wash Diluent (DIL) a temperatura ambiente.
Mezclar el contenido del vial Wash Diluent (DIL) minuciosamente mediante vórtex, centrifugar brevemente y colocar en hielo.
En un tubo Eppendorf DNA LoBind nuevo de 1,5 ml preparar la siguiente mezcla de lavado de la celda de flujo:
| Reactivo | Volumen por celda de flujo |
|---|---|
| Wash Mix (WMX) | 2 μl |
| Wash Diluent (DIL) | 398 μl |
| Total | 400 μl |
Mezclar bien con la pipeta y colocar en hielo. No mezclar el tubo en vórtex.
Detener el experimento de secuenciación en MinKNOW y dejar la celda de flujo en el dispositivo.
Es fundamental que el puerto de entrada esté cerrado antes de eliminar los desechos para evitar que el aire atraviese la zona de la matriz de sensores, que provocaría una pérdida significativa de canales de secuenciación.
Retirar el tampón de desecho de la siguiente manera:
- Cerrar el puerto de entrada.
- Insertar una pipeta P1000 en el puerto de desecho y retirar el tampón de desecho.
Nota: Como el orificio de entrada está cerrado, ningún fluido debe salir de la zona de la matriz de sensores.
Deslizar la tapa del puerto de entrada en el sentido de las agujas del reloj.
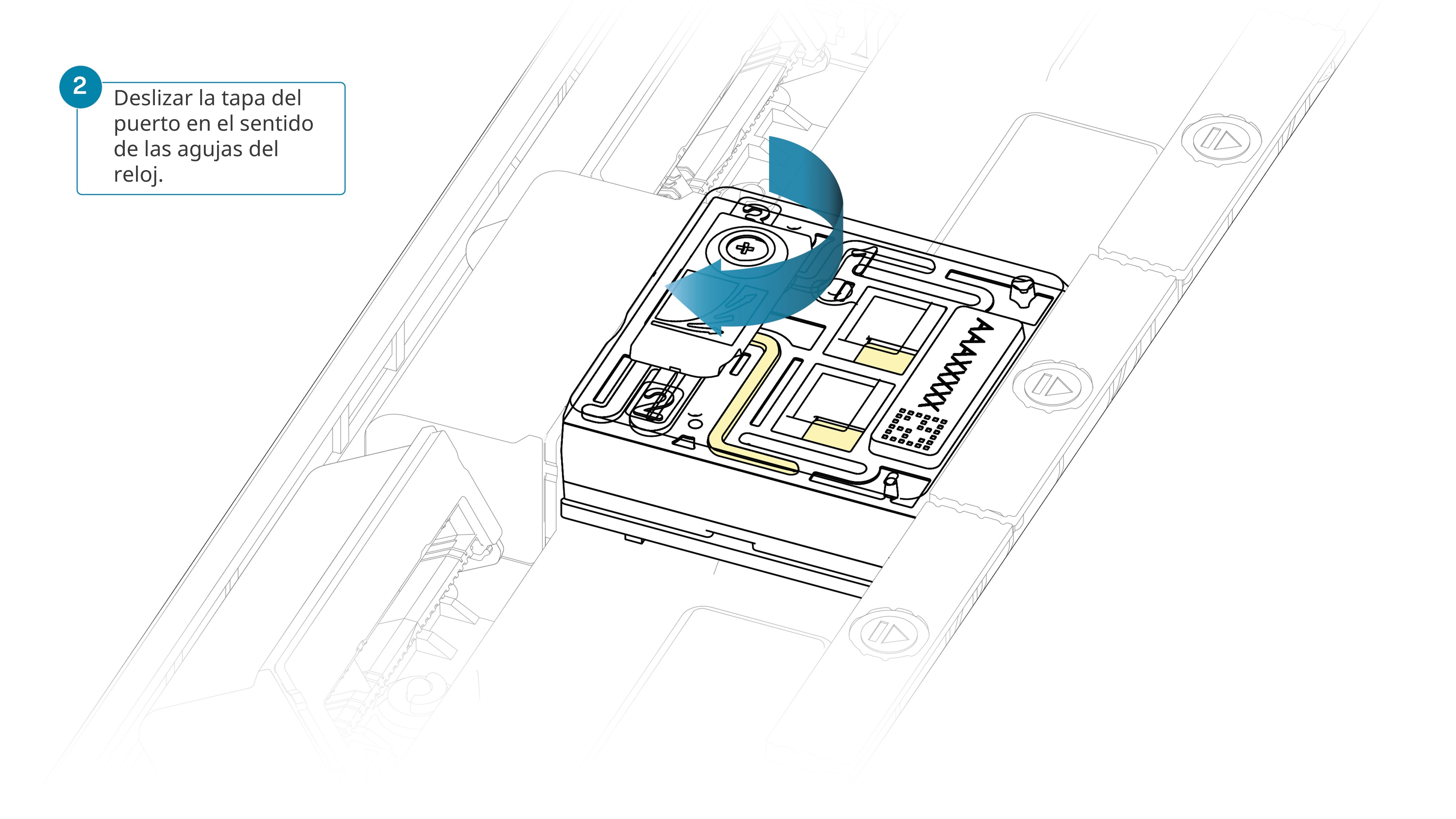
Tenga cuidado al extraer tampón de la celda de flujo. No retire más de 20-30 μl y compruebe que el tampón cubra la matriz de poros en todo momento. La introducción de burbujas de aire en la matriz puede dañar los poros de manera irreversible.
Tras abrir el puerto de entrada, comprobar si hay burbujas de aire bajo la tapa. Retirar una pequeña cantidad de fluido para quitar las burbujas:
- Ajustar una pipeta P1000 a 200 μl.
- Introducir la punta de la pipeta en el puerto de entrada.
- Girar la rueda hasta que el indicador de volumen marque 220-230 μl o hasta que se pueda ver una pequeña cantidad entrar en la punta de la pipeta.

Cargar 200 μl de mezcla de lavado por el puerto de entrada, de la siguiente manera:
- Con una pipeta P1000, extraer 200 µl de mezcla de lavado de la celda de flujo
- Introducir la punta de la pipeta en el puerto de entrada, procurando que no haya burbujas de aire en la punta.
- Girar lentamente la rueda de la pipeta hacia abajo (si es posible) para cargar la celda de flujo o empujar muy lentamente el émbolo hacia abajo y dejar una cantidad pequeña en la punta de la pipeta.
- Programar el temporizador e incubar durante 5 minutos.
Una vez acabado el tiempo de incubación, cargar con cuidado los 200 μl de mezcla de lavado restante en el puerto de entrada, de la siguiente manera:
- Con una pipeta P1000, extraer 200 µl de mezcla de lavado de la celda de flujo.
- Introducir la punta de la pipeta en el puerto de entrada, procurando que no haya burbujas de aire en la punta.
- Girar lentamente la rueda de la pipeta hacia abajo (si es posible) para cargar la celda de flujo o empujar muy lentamente el émbolo hacia abajo y dejar una cantidad pequeña en la punta de la pipeta.
Cerrar el puerto de entrada y esperar 1 hora.

Es fundamental que el puerto de entrada esté cerrado antes de eliminar los desechos para evitar que el aire atraviese la zona de la matriz de sensores, lo que provocaría una pérdida significativa de canales de secuenciación.
Retirar el tampón de desecho de la siguiente manera:
- Compruebe que el puerto de entrada esté cerrado.
- Introducir una pipeta P1000 en el puerto de desecho y retirar el tampón.
Nota: como el puerto de entrada está cerrado, ningún fluido debe salir del área de la matriz de sensores.
Los tampones utilizados en este proceso son incompatibles con verificar la celda de flujo antes de cargar la biblioteca siguiente. Sin embargo, el número de poros disponible se comunicará después de la siguiente exploración de poros.
Descongelar los viales Sequencing Buffer (SB), Library Beads (LIB) o Library Solution (LIS), -si se requiere-, Flow Cell Tether (FCT) y un tubo de Flow Cell Flush (FCF) a temperatura ambiente y mezclar en vórtex. Centrifugar brevemente y poner en hielo.
Preparar la mezcla de acondicionamiento en un tubo adecuado al número de celdas de flujo que purgar. Una vez combinado, mezclar bien mediante un breve vórtex.
| Reactivo | Volumen por celda de flujo |
|---|---|
| Flow Cell Flush (FCF) | 1 170 µl |
| Flow Cell Tether (FCT) | 30 µl |
| Total volume | 1 200 µl |
Deslizar el puerto de entrada en el sentido de las agujas del reloj.

Tenga cuidado al extraer tampón de la celda de flujo. No retire más de 20-30 μl y compruebe que el tampón cubra la matriz de poros en todo momento. La introducción de burbujas de aire en la matriz puede dañar los poros de manera irreversible.
Tras abrir el puerto de entrada, retirar una pequeña cantidad de tampón para quitar las burbujas de aire:
- Ajustar una pipeta P1000 a 200 μl.
- Introducir la punta de la pipeta en el puerto de entrada.
- Girar la rueda hasta que el indicador de volumen marque 220-230 μl o hasta que se pueda ver una pequeña cantidad entrar en la punta de la pipeta.

Cargar 500 μl de mezcla de acondicionamiento por el puerto de entrada, de la siguiente manera:
- Con una pipeta P1000, extraer 500 µl de mezcla de acondicionamiento
- Introducir la punta de la pipeta en el puerto de entrada, procurando que no haya burbujas de aire en la punta.
- Girar lentamente la rueda de la pipeta hacia abajo (si es posible) para cargar la celda de flujo o empujar muy lentamente el émbolo hacia abajo y dejar una cantidad pequeña en la punta de la pipeta.

Es esencial esperar cinco minutos entre los purgados con mezcla de acondicionamiento para garantizar la eliminación eficaz de la nucleasa.
Cerrar el puerto de entrada y esperar cinco minutos.
Durante este tiempo, preparar la biblioteca para cargar siguiendo los siguientes pasos del protocolo.
Mezclar minuciosamente con la pipeta el contenido del vial Library Beads (LIB).
El vial Library Beads (LIB) contiene microesferas en suspensión. Las microesferas sedimentan muy rápido; por eso es crucial mezclarlas justo antes de su uso.
Recomendamos utilizar Library Beads (LIB) en la mayoría de experimentos de secuenciación. No obstante, el reactivo Library Solution (LIS) está disponible en caso de utilizar bibliotecas más viscosas.
En un tubo Eppendorf DNA LoBind de 1,5 ml preparar la biblioteca de la siguiente manera:
| Reactivo | Volumen por celda de flujo |
|---|---|
| Sequencing Buffer (SB) | 100 µl |
| Library Beads (LIB) bien mezclado antes de su uso | 68 µl |
| Biblioteca de ADN | 32 µl |
| Total | 200 µl |
Nota: hemos aumentado el volumen de carga de la biblioteca, para mejorar la cobertura de la matriz.
Es fundamental que el puerto de entrada esté cerrado antes de eliminar los desechos para evitar que el aire atraviese la zona de la matriz de sensores, lo que provocaría una pérdida significativa de canales de secuenciación.
Retirar el tampón de desecho de la siguiente manera:
- Compruebe que el puerto de entrada esté cerrado.
- Introducir una pipeta P1000 en el puerto de desecho y retirar el tampón.
Nota: como el puerto de entrada está cerrado, ningún fluido debe salir del área de la matriz de sensores.
Deslizar la tapa del puerto de entrada en el sentido de las agujas del reloj.

Tenga cuidado al extraer tampón de la celda de flujo. No retire más de 20-30 μl y compruebe que el tampón cubra la matriz de poros en todo momento. La introducción de burbujas de aire en la matriz puede dañar los poros de manera irreversible.
Tras abrir el puerto de entrada, retirar una pequeña cantidad de tampón para quitar las burbujas de aire:
- Ajustar una pipeta P1000 a 200 μl.
- Introducir la punta de la pipeta en el puerto de entrada.
- Girar la rueda hasta que el indicador de volumen marque 220-230 μl o hasta que se pueda ver una pequeña cantidad entrar en la punta de la pipeta.

Cargar 500 μl de mezcla de acondicionamiento por el puerto de entrada, de la siguiente manera:
- Con una pipeta P1000, extraer 500 µl de mezcla de acondicionamiento
- Introducir la punta de la pipeta en el puerto de entrada, procurando que no haya burbujas de aire en la punta.
- Girar lentamente la rueda de la pipeta hacia abajo (si es posible) para cargar la celda de flujo o empujar muy lentamente el émbolo hacia abajo y dejar una cantidad pequeña en la punta de la pipeta.

Es fundamental que el puerto de entrada esté cerrado antes de eliminar los desechos para evitar que el aire atraviese la zona de la matriz de sensores, lo que provocaría una pérdida significativa de canales de secuenciación.
Retirar el tampón de desecho de la siguiente manera:
- Cerrar el puerto de entrada.
- Insertar una pipeta P1000 en el puerto de desecho y retirar el tampón.
Nota: como el orificio de entrada está cerrado, ningún fluido debe salir de la zona de la matriz de sensores.
Mover la tapa del puerto de entrada en el sentido de las agujas del reloj.

Tenga cuidado al extraer tampón de la celda de flujo. No retire más de 20-30 μl y compruebe que el tampón cubra la matriz de poros en todo momento. La introducción de burbujas de aire en la matriz puede dañar los poros de manera irreversible.
Tras abrir el puerto de entrada, retirar una pequeña cantidad de tampón para quitar las burbujas de aire:
- Ajustar una pipeta P1000 a 200 μl.
- Introducir la punta de la pipeta en el puerto de entrada.
- Girar la rueda hasta que el indicador de volumen marque 220-230 μl o hasta que se pueda ver una pequeña cantidad entrar en la punta de la pipeta.

Mezclar la biblioteca suavemente con la pipeta antes de cargarla.
Cargar 200 μl de biblioteca con una pipeta p1000.

Cerrar la válvula, lo que sellará el puerto de entrada.

A fin de lograr un rendimiento de secuenciación óptimo, colocar la pantalla protectora sobre la celda de flujo tan pronto como se haya cargado la biblioteca.
Recomendamos dejar la pantalla protectora colocada mientras la celda de flujo esté cargada, también durante los lavados o recargas. La pantalla puede quitarse una vez que se haya extraído la biblioteca de la celda de flujo.
Si se ha retirado la pantalla protectora de la celda de flujo, se instala de la siguiente manera:
- Alinear la abertura de la pantalla protectora con la tapa del puerto de entrada de la celda de flujo. El borde delantero de la pantalla protectora debe quedar por encima del identificador de la celda de flujo.
- Presionar la pantalla protectora con firmeza alrededor de la tapa del puerto de entrada. Cuando el clip del puerto de entrada encaje bajo la tapa del mismo, se producirá un clic.


Cerrar la tapa del PromethION cuando esté listo para empezar un experimento de secuenciación en MinKNOW.
Esperar al menos 10 minutos tras cargar las celdas de flujo en el PromethION antes de iniciar cualquier experimento ayudará a aumentar el rendimiento de la secuenciación.
Realizar el paso «Lavado y recarga de celdas de flujo PromethION» dos veces, de modo que se hagan tres cargas en total: la carga de la biblioteca inicial más dos lavados y recargas. El objetivo es maximizar la adquisición de datos.
- El primer lavado y recarga debe realizarse tras 24 h de secuenciación aproximadamente.
- El segundo lavado y recarga debe realizarse tras 48 h de secuenciación aproximadamente.
11. Adquisición de datos e identificación de bases
Aspectos generales del análisis de datos de nanoporos
Encontrará una descripción completa del análisis de datos de nanoporos, con distintas posibilidades de análisis de identificación y postidentificicación de bases, en el documento Data Analysis.
Cómo empezar a secuenciar
El programa MinKNOW gestiona el dispositivo de secuenciación y la adquisición de datos. Compruebe que MinKNOW esté instalado en su ordenador. Encontrará instrucciones adicionales para configurar experimentos de secuenciación en el Protocolo de MinKNOW.
Ajustes de secuenciación en el protocolo de secuenciación múltiple de metilación con representación reducida:
En la pestaña del kit, seleccionar Native Barcoding Sequencing Kit 24 (SQK-NBD114.24).
DESACTIVAR la identificación de bases (que desactivará automáticamente la adición de códigos de barras).
Nota: la identificación de bases y adición de códigos se llevará a cabo tras la secuenciación, en la sección de análisis de datos.ACTIVAR el Muestreo adaptativo y seleccionar Enriquecer.
Introducir el archivo de referencia humano para la alineación y el archivo .bed para enumerar las regiones (consulte el catálogo en línea del archivo .bed de RRMS humano).Configurar la duración del experimento a un mínimo de 96 horas.
Establecer los parámetros de salida deseados.
Recomendamos mantener las opciones predeterminadas de formato (.POD5) en el archivo de salida, lo que garantiza el correcto funcionamiento del análisis posterior.Pulsar Empezar.
12. Análisis de datos
Versiones de programas informáticos
A continuación, están las versiones de programas informáticos utilizadas en esta guía. Tenga en cuenta que las versiones más recientes del programa tal vez no sean compatibles con algunos comandos incluidos en esta guía.
| Programas informáticos | Versión |
|---|---|
| dorado | v0.7.3 |
| modkit | v0.2.8 |
| wf-human-variation | v2.3.0 |
| mosdepth | v0.3.8 |
Identificación de bases y desmultiplexado:
Identificación de bases
La versión independiente de Dorado se utiliza para realizar la identificación de bases. Abrir una ventana del terminal e introducir los siguientes comandos:
dorado basecaller hac, 5mCG_5hmCG --kit-name SQK-NBD114-24 \
--secondary “no” -Y \
--reference {reference_fasta} {input_pod5_folder} \
| samtools view -e '[qs] >= {qscore_filter}' \
--output {out_pass_bam} \
--unoutput {out_fail_bam}
Notas:
- Recomendamos utilizar el modelo de gran precisión (HAC), en los ciclos de secuenciación de RRMS. No obstante, si utiliza el modelo de ultra precisión (SUP), compruebe que utiliza el modelo correcto en los comandos mencionados arriba.
- Es posible realizar la alineación durante la identificación de bases, con ayuda de un archivo FASTA. El archivo de referencia humana recomendado se encuentra disponible para descargar aquí.
- Las alineaciones secundarias se descartan mediante la opción «--secondary no» y se activa la opción Y, lo que permite un suave corte de alineaciones complementarias.
- Al proporcionar la opción «--kit-name SQK-NBD114.24», Dorado también clasificará las lecturas entre los diferentes códigos de barras presentes, con una etiqueta en el archivo BAM generado.
- Recomendamos ajustar el filtro Q score a 10.
- Tenga en cuenta, para identificar bases con Dorado es necesario el procesamiento en GPU. Encontrará más información sobre cómo ejecutar Dorado en el repositorio de GitHub.
Desmultiplexado
Dorado demux se utiliza para clasificar lecturas por código de barras mediante el siguiente comando:
dorado demux --no-classify --sort-bam --output-dir <out_folder> {out_pass_bam}
Notas:
- Este paso generará archivos BAM separados por cada uno de los códigos de barras posibles relacionados con el kit utilizado (por ejemplo, SQK-NBD114-24).
- El recorte de adaptadores y códigos de barras se realizará por defecto en Dorado. Una vez recortados los códigos de barras, no es posible desmultiplexar las lecturas de nuevo.
- Encontrará más información en la documentación de Dorado.
Análisis de la cobertura:
El archivo .bed con las regiones utilizadas en RRMS se encuentra disponible en el catálogo AS.
Mosdepth se utiliza para comprobar la cobertura en regiones seleccionadas de los códigos de barras de interés:
mosdepth -x -t 8 -n -b {target_bed} {out_prefix} {input_pass_bam}
Identificación de las modificaciones:
El flujo bioinformático de variación humana se utiliza para agregar modificaciones en posiciones genómicas mediante modkit.
El flujo de trabajo está disponible en el siguiente repositorio: wf-human-variation github.
La documentación se encuentra en el siguiente espacio: wf-human-variation EPI2ME page
En la mayoría de los experimentos de RRMS recomendamos utilizar el siguiente comando:
nextflow run https://github.com/epi2me-labs/wf-human-variation \
-profile singularity \
--mod \
--bam <bam> \
--bed RRMS_human_hg38.bed \
--ref GCA_000001405.15_GRCh38_no_alt_analysis_set.fasta \
--sample_name <sample> --out_dir <output_dir>
(Opcional) En la metilación específica del haplotipo
Si se necesita metilación específica del haplotipo, es posible proporcionar opciones «--snp – phased» para agregar modificaciones identificadas en cada uno de los haplotipos (es decir, se generará un archivo bedmethyl de metilación para cada uno de los haplotipos):
nextflow run https://github.com/epi2me-labs/wf-human-variation \
-profile singularity \
--mod --snp --phased \
--bam <bam> \
--bed RRMS_human_hg38.bed \
--ref GCA_000001405.15_GRCh38_no_alt_analysis_set.fasta \
--sample_name <sample> --out_dir <output_dir>
Nota: en este análisis específico, recomendamos una cobertura de muestra superior a 30×.
Detección de regiones diferencialmente metiladas:
«modkit dmr» sirve para detectar regiones diferencialmente metiladas a través de diferentes muestras.
Encontrará más información en la documentación de modkit.
Visualización:
Los archivos BAM generados por Dorado contienen bases canónicas así como modificaciones por lectura guardadas en etiquetas MM y ML (del archivo BAM). Para visualizar las modificaciones por lectura en IGV (Integrative Genomics Viewer), cargue el archivo BAM y seleccione «base modification 2-color (all)» en el menú «colour reads as».
Si se ha realizado la asignación de variantes a haplotipos mediante el flujo bioinformático wf-human-variation, es posible cargar el archivo BAM con etiquetas haplotípicas en IGV y agrupar las alineaciones por haplotipo mediante la opción «group by» y la selección de «phase».
En IGV también es posible visualizar las frecuencias de metilación por posición mediante el formato BIGWIG. Para ello, se utiliza modkit, que genera archivos BEDGRAPH mediante el siguiente comando:
modkit pileup --cpg --combine-strands --bedgraph \
--threads 10 --prefix {out_prefix} \
--ref {reference_fasta} \
{out_folder} {input_pass_bam}
Tenga en cuenta que se creará un archivo BEDGRAPH diferente para cada una de las modificaciones presentes, en este caso 5mC y 5hmC.
BedGraphToBigWig se utiliza para generar archivos bigwig, que es posible cargar en IGV junto con el archivo BAM:
bedtools sort -i {out_folder}/{prefix}_m_CG0_combined.bedgraph | cut -f 1-4 > {out_folder}/{prefix}_m_CG0_combined_sort.bedgraph
bedGraphToBigWig {out_folder}/{prefix}_m_CG0_combined_sort.bedgraph {reference_chrSize} {out_mod_bed_agg_filt_bigwig}
Resultados de la evaluación comparativa
Encontrará información sobre la evaluación comparativa del rendimiento de RRMS en muestras humanas, en el documento de rendimiento RRMS
13. Reutilización y retorno de las celdas de flujo
Con este método, no recomendamos lavar y reutilizar las celdas de flujo.
Debido al prolongado tiempo de secuenciación y a los múltiples lavados de las celdas de flujo y recargas de bibliotecas, no recomendamos reutilizar las celdas de flujo utilizadas en este protocolo.
Reutilizar las celdas de flujo en experimentos posteriores tal vez dé como resultado que se generen datos insuficientes para el análisis.
Siga el procedimiento de devolución y envíe las celdas de flujo a Oxford Nanopore para que las recicle.
Aquí encontrará las instrucciones para devolver celdas de flujo.
Ante cualquier problema o pregunta acerca del experimento de secuenciación, consulte la sección Resolución de problemas de la versión en línea de este protocolo.
14. Problemas durante la extracción de ADN y la preparación de la biblioteca
A continuación, hay una lista de los problemas más frecuentes, con algunas posibles causas y soluciones propuestas.
También disponemos de una sección de preguntas frecuentes, FAQ, en la sección Support de la comunidad Nanopore.
Si ha probado las soluciones propuestas y los problemas aún persisten, póngase en contacto con el departamento de Asistencia técnica, bien por correo electrónico (support@nanoporetech.com) o a través del chat en directo de la comunidad Nanopore.
Baja calidad de la muestra
| Observaciones | Posibles causas | Comentarios y acciones recomendadas |
|---|---|---|
| Baja pureza del ADN (la lectura del Nanodrop para ADN OD 260/280 es <1,8 y OD 260/230 es <2,0-2,2) | El método de extracción de ADN no proporciona la pureza necesaria | Los efectos de los contaminantes se muestran en el documento Contaminants. Probar con otro método de extracción que no provoque el arrastre de contaminantes. Considere realizar un paso adicional de limpieza SPRI. |
Escasa recuperación de ADN tras la limpieza con microesferas AMPure
| Observaciones | Posibles causas | Comentarios y acciones recomendadas |
|---|---|---|
| Escasa recuperación | Pérdida de ADN debido a una proporción de microesferas AMPure por muestra inferior a lo previsto. | 1. Las microesferas AMPure sedimentan con rapidez; antes de añadirlas a la muestra deben estar bien resuspendidas. 2. Si la proporción de microesferas AMPure por muestra es inferior a 0.4:1, los fragmentos de ADN, sean del tamaño que sean, se perderán durante la limpieza. |
| Escasa recuperación | Los fragmentos de ADN son más cortos de lo esperado | Cuanto menor sea la proporción de microesferas AMPure por muestra, más rigurosa será la selección de fragmentos largos frente a los cortos. Determinar siempre la longitud de la muestra de ADN en un gel de agarosa u otros métodos de electroforesis en gel y a continuación, calcular la cantidad adecuada de microesferas AMPure que se debe utilizar. 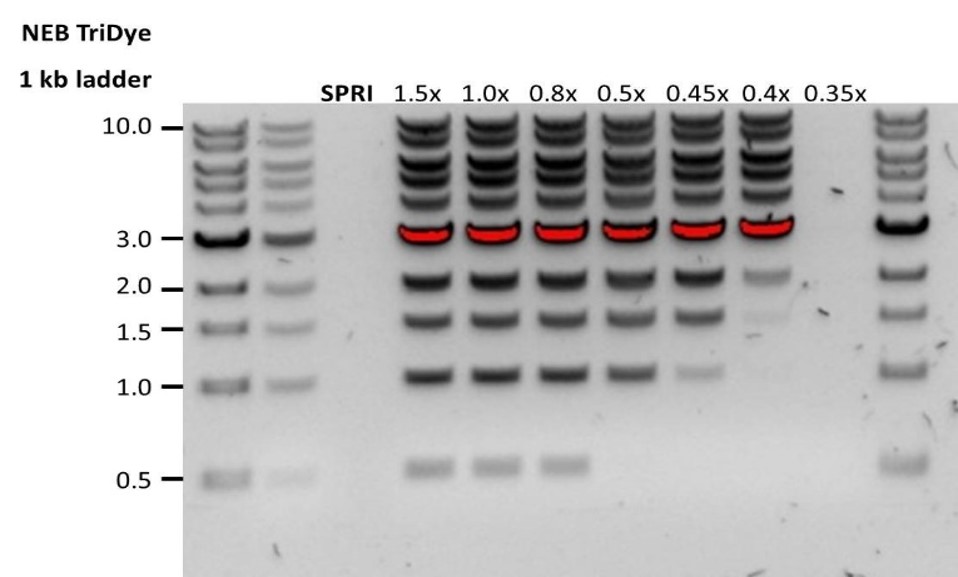 |
| Escasa recuperación tras la preparación de extremos | En el paso de lavado se utilizó etanol en una concentración inferior al 70 % | Si se utiliza etanol a una concentración inferior al 70 %, el ADN se eluye de las microesferas. No olvide utilizar el porcentaje correcto. |
15. Problemas durante el experimento de secuenciación
A continuación hay una lista de los problemas más frecuentes, con algunas posibles causas y soluciones propuestas.
También disponemos de una sección de preguntas frecuentes, FAQ, en la sección Support de la comunidad Nanopore.
Si ha probado las soluciones propuestas y los problemas aún persisten, póngase en contacto con el departamento de Asistencia técnica, bien por correo electrónico (support@nanoporetech.com) o a través del chat Live de la comunidad Nanopore.
Menos poros al inicio de la secuenciación que tras la evaluación de la celda de flujo
| Observaciones | Posibles causas | Comentarios y acciones recomendadas |
|---|---|---|
| MinKNOW presentó al inicio de la secuenciación un número de poros inferior al indicado durante la evaluación de la celda de flujo. | Se introdujo una burbuja de aire en la matriz de nanoporos. | Tras comprobar el número de poros presentes en la celda de flujo y antes de acondicionarla, es imprescindible quitar las burbujas de aire que haya cerca del puerto de purgado. Si no se quitan, las burbujas de aire pueden desplazarse a la matriz de nanoporos y dañar de manera irreversible los nanoporos expuestos al aire. En [este vídeo] (https://nanoporetech.com/resource-centre/masterclass-how-to-load-a-promethion-flow-cell-lc25) se muestran algunas buenas prácticas para evitar que esto ocurra. |
| MinKNOW presentó al inicio de la secuenciación un número de poros inferior al indicado durante la evaluación de la celda de flujo | La celda de flujo no está colocada correctamente. | Detener el ciclo de secuenciación, quitar la celda de flujo del dispositivo e insertarla de nuevo. Comprobar que está firmemente asentada en el dispositivo y que ha alcanzado la temperatura deseada. Si procede, probar con una posición diferente del dispositivo (GriION/PromethION). |
| MinKNOW presentó al inicio de la secuenciación un número de poros inferior al indicado durante la evaluación de la celda de flujo | La presencia de contaminantes en la biblioteca ha dañado o bloqueado los poros. | El recuento de poros durante la evaluación de la celda de flujo se efectúa utilizando las moléculas de ADN del control de calidad presentes en el tampón de almacenamiento. Al inicio de la secuenciación, se utiliza la misma biblioteca para calcular el número de poros activos. Por este motivo, se estima que puede haber una variabilidad del 10 % en el número de poros detectados. Tener un número de poros considerablemente inferior al inicio de la secuenciación puede deberse a la presencia de contaminantes en la biblioteca que hayan dañado las membranas o bloqueado los poros. Para mejorar la pureza del material de entrada tal vez sea necesario usar métodos de purificación o extracción de ADN/ARN alternativos. Los efectos de los contaminantes se muestran en la página Contaminants. Probar con un método de extracción diferente que no provoque el arrastre de contaminantes. |
Error en el script de MinKNOW
| Observaciones | Posibles causas | Comentarios y acciones recomendadas |
|---|---|---|
| MinKNOW muestra el mensaje "Error en el script" | Reiniciar el ordenador y reiniciar MinKNOW. Si el problema persiste, reúna los archivos de registro MinKNOW log files y contacte con el departamento de Asistencia técnica. Si no dispone de otro dispositivo de secuenciación, recomendamos que guarde la celda de flujo cargada a 4 °C y contacte con el servicio de asistencia técnica para recibir instrucciones de almacenamiento adicionales. |
Ocupación de poro por debajo del 40 %
| Observaciones | Posibles causas | Comentarios y acciones recomendadas |
|---|---|---|
| Ocupación de poro <40 % | No se cargó suficiente cantidad de biblioteca en la celda de flujo | Cargue la biblioteca al volumen y la concentración adecuados, tal como se indica en el protocolo correspondiente. Cuantificar la biblioteca antes de cargarla y calcular fmoles con herramientas como la calculadora Biomath de Promega, y escoger la opción "dsDNA: μg to fmol". |
| Ocupación de poro próxima a 0 | Se utilizó el kit Ligation Sequencing Kit y los adaptadores de secuenciación no se ligaron al ADN | En la fase de ligación del adaptador, añadir la cantidad recomendada de NEBNext Quick Ligation Module (E6056) y el tampón Ligation Buffer (LNB) de Oxford Nanopore Technologies, suministrado en el kit de secuenciación. Preparar una biblioteca de control con lambda para valorar la integridad de los reactivos de otros fabricantes. |
| Ocupación de poro próxima a 0 | Se utilizó el kit Ligation Sequencing Kit y en la fase de lavado, después de la ligación del adaptador, se utilizó etanol en lugar de Long Fragment Buffer (LFB) o Short Fragment Buffer (SFB). | El etanol puede desnaturalizar la proteína motor en los adaptadores de secuenciación. Usar Long Fragment Buffer (LFB) o Short Fragment Buffer (SFB) después de la ligación de los adaptadores. |
| Ocupación de poro próxima a 0 | No hay anclaje en la celda de flujo | Los anclajes se añaden durante el acondicionamiento de la celda de flujo (viales (FLT)/(FCT)). Añadir Flush Tether (FLT)/Flow Cell Tether (FCT) al vial Flush Buffer (FB)/Flow cCell Flush (FCF) antes de acondicionar la celda de flujo. |
Longitud de lectura más corta de lo esperado
| Observaciones | Posibles causas | Comentarios y acciones recomendadas |
|---|---|---|
| Longitud de lectura más corta de lo esperado | Fragmentación no deseada de la muestra de ADN | La longitud de lectura refleja la longitud del fragmento de la muestra de ADN. La muestra de ADN se puede fragmentar durante la extracción y la preparación de la biblioteca. 1. Consulte la sección de buenas prácticas de extracción en la página Extraction Methods de la comunidad Nanopore. 2. Visualizar la distribución de la longitud de los fragmentos de las muestras de ADN en un gel de agarosa antes de proceder a la preparación de la biblioteca.  En la imagen superior, la muestra 1 contiene un alto peso molecular, mientras que la muestra 2 se ha fragmentado. En la imagen superior, la muestra 1 contiene un alto peso molecular, mientras que la muestra 2 se ha fragmentado.3. Durante la preparación de la biblioteca, evitar pipetear y agitar en vórtex al mezclar los reactivos. Dar suaves golpes con el dedo o invertir el vial es suficiente. |
Gran proporción de poros no disponibles
| Observaciones | Posibles causas | Comentarios y acciones recomendadas |
|---|---|---|
Gran proporción de poros no disponibles (se muestran en azul en el panel de canales y en el gráfico de actividad de poros) 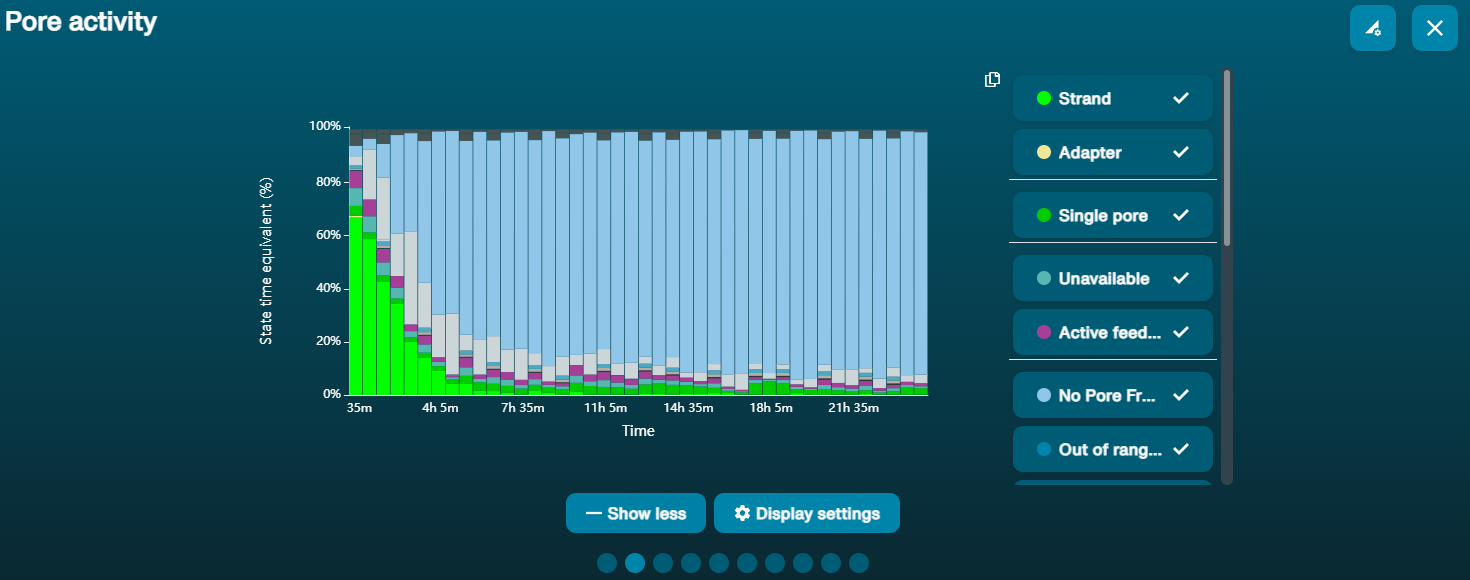 El gráfico de actividad de poros de arriba muestra una proporción creciente de poros "no disponibles» conforme pasa el tiempo. El gráfico de actividad de poros de arriba muestra una proporción creciente de poros "no disponibles» conforme pasa el tiempo. | Hay contaminantes presentes en la muestra | Algunos contaminantes se pueden eliminar de los poros mediante la función de desbloqueo incorporada en MinKNOW. Si funciona, el estado de los poros cambiará a "sequencing pores". Si la porción poros no disponibles se mantiene elevada o aumenta: 1. Realizar un purgado de nucleasa con el kit de lavado Flow Cell Wash Kit (EXP-WSH004) o 2. Realizar varios ciclos de PCR para intentar diluir cualquier contaminante que pueda estar causando problemas. |
Gran proporción de poros inactivos
| Observaciones | Posibles causas | Comentarios y acciones recomendadas |
|---|---|---|
| Gran proporción de poros inactivos/no disponibles (se muestran en azul claro en el panel de canales y en el gráfico de actividad de poros. Los poros o membranas están dañados de manera irreversible) | Se han introducido burbujas de aire en la celda de flujo | Las burbujas de aire introducidas durante el acondicionamiento de la celda de flujo y carga de la biblioteca podrían dañar los poros de forma permanente. La forma más eficaz de evitar que esto ocurra se muestra en el vídeo Como cargar celdas de flujo. |
| Gran proporción de poros inactivos/no disponibles | Ciertos compuestos copurificados con ADN | Algunos compuestos conocidos, incluidos los polisacáridos. 1. Purificar usando el kit PowerClean Pro de QIAGEN. 2. Realizar una amplificación del genoma completo con la muestra original de ADNg utilizando el kit REPLI-g de QIAGEN. |
| Gran proporción de poros inactivos/no disponibles | Hay contaminantes presentes en la muestra | Los efectos de los contaminantes se muestran en la página Contaminants. Probar con un método de extracción alternativo que no provoque el arrastre de contaminantes. |
Fluctuación de la temperatura
| Observaciones | Posibles causas | Comentarios y acciones recomendadas |
|---|---|---|
| Fluctuación de la temperatura | La celda de flujo ha perdido contacto con el dispositivo | Comprobar que una almohadilla térmica cubra la placa metálica de la parte posterior de la celda de flujo. Reinsertarla y presionar hasta que las clavijas del conector estén firmemente en contacto con el dispositivo. Si el problema persiste, contacte con el servicio de Asistencia técnica. |
Error al intentar alcanzar la temperatura deseada
| Observaciones | Posibles causas | Comentarios y acciones recomendadas |
|---|---|---|
| MinKNOW muestra el mensaje "Error al intentar alcanzar la temperatura deseada" | El dispositivo ha sido colocado en un lugar con una temperatura ambiente inferior a la media o en un lugar con escasa ventilación (lo que provoca el sobrecalientamiento de las celdas de flujo). | MinKNOW dispone de un tiempo predeterminado para que las celdas de flujo alcancen la temperatura fijada. Una vez acabado el tiempo, aparece un mensaje de error, pero el experimento de secuenciación continúa. Secuenciar a una temperatura incorrecta puede llevar a una disminución en el rendimiento y generar índices de calidad Qscore menores. Corregir la ubicación del dispositivo, procurando que esté a temperatura ambiente y con buena ventilación; a continuación, reiniciar el proceso en MinKNOW. |







