Telomere-to-telomere sequencing (T2T) on PromethION (SQK-LSK114-XL and SQK-ULK114) (T2T_9179_v114_revL_31Jul2024)
PromethION: Protocol
V T2T_9179_v114_revL_31Jul2024
FOR RESEARCH USE ONLY
Contents
Introduction to the protocol
Reagent and sample preparation
- 4. Custom SPRI bead preparation for the Pore-C experiment
- 5. Whole blood sample preparation for the Pore-C experiment
- 6. Whole blood sample gDNA extraction for the Duplex experiment
- 7. Whole blood sample preparation for the Ultra-long DNA experiment
Day 1: Library preparation
- 8. Day 1: Pore-C experiment
- 9. Day 1: Duplex experiment
- 10. Day 1: Priming and loading Duplex library on the PromethION Flow Cell
Day 2: Library preparation
- 11. Day 2: Pore-C experiment
- 12. Day 2: Ultra-long DNA experiment
- 13. Day 2: Washing and reloading Duplex library on the PromethION Flow Cell
Day 3: Library preparation
- 14. Day 3: Pore-C experiment
- 15. Day 3: Ultra-long DNA experiment
- 16. Day 3: Priming and loading ultra-long DNA library on the PromethION Flow Cell
- 17. Day 3: Washing and reloading Duplex library on the PromethION Flow Cell
Day 4: Library preparation
- 18. Day 4: Pore-C experiment
- 19. Day 4: Priming and loading Pore-C library on the PromethION Flow Cell
- 20. Day 4: Washing and reloading Duplex library on the PromethION Flow Cell
- 21. Day 4: Washing and reloading the PromethION Flow Cell with ultra-long DNA library
Day 5: Wash and reload flow cells
- 22. Day 5: Washing and reloading Pore-C library on the PromethION Flow Cell
- 23. Day 5: Washing and reloading the PromethION Flow Cell with ultra-long DNA library
Day 6: Wash and reload flow cells
Day 7: Wash and reload flow cells
Sequencing and data analysis
- 26. Duplex experiment: Data acquisition and basecalling
- 27. Ultra-long DNA experiment: Data acquisition and basecalling
- 28. Pore-C experiment: Data acquisition and basecalling
- 29. Downstream analysis
- 30. Flow cell reuse and returns
Troubleshooting
1. Overview of the protocol
This is a Legacy protocol
Please use our new Telomere-to-telomere sequencing (T2T) on PromethION (SQK-APK114, SQK-LSK114, and SQK-ULK114) protocol for this application.
For more information on Nanopore-only Telomere-to-telomere (T2T) or to register your interest please follow this link.
For more information about our Early Access programmes, please see this article on product release phases.
Introduction to the protocol
This protocol describes an end-to-end workflow for telomere-to-telomere sequencing of the human genome using the Oxford Nanopore PromethION platform. The protocol includes three separate sequencing experiments, each requiring at least one PromethION Flow Cell, which are carried out over seven days. The library preparation, sequencing and flow cell washing and reloading steps are described day-by-day.
This protocol was developed in collaboration with the UCSC Nanopore Production Center, led by Dr. Karen Miga.
To generate as much data as possible, multiple flow cell washes are required to recover pores and reload fresh library to maximise data output. Depending on sample quality and DNA yield post-library preparation, the library preparation steps may need to be scaled up to meet the number of fresh libraries required for each flow cell wash and library reload. Excess DNA libraries can be stored in Eppendorf DNA LoBind tubes at 4°C until loading on the required day.
Three different datasets are generated to give both high accuracy duplex data and ultra-long reads, alongside the chromatin conformation capture data to achieve in-depth telomere-to-telomere sequencing of a sample.
Three experiments are set up across seven days:
Duplex experiment: This experiment generates high accuracy duplex data using the Ligation Sequencing Kit XL V14 (SQK-LSK114-XL). Library preparation takes ~60 minutes hands-on time, followed by three washes across a 100-hour sequencing run.
Ultra-long DNA experiment: This experiment generates ultra-long reads using the Ultra-Long DNA Sequencing Kit V14 (SQK-ULK114). DNA extraction and library preparation takes ~3.5 hours with an overnight elution, followed by two washes across a 72-hour sequencing run. This experiment generates a very viscous library of very long DNA fragments which requires careful handling to maintain the long fragments.
Pore-C experiment: This experiment produces chromatin conformation capture data using the Pore-C protocol and the Ligation Sequencing Kit XL V14 (SQK-LSK114-XL). The Pore-C DNA extraction takes ~3 hours hands-on time over three days with two overnight steps. The library preparation takes ~60 minutes hands-on time, followed by three washes across a 72-hour sequencing run. This experiment has been developed by Oxford Nanopore Technologies and the following published literature: Lieberman-Aiden et al., 2009; Comet et al., 2011; Belton et al., 2012; Gavrilov, Golov and Razin, 2013; Nagano et al., 2015; Belaghzal, Dekker and Gibcus, 2017; Ulahannan et al., 2019. This experiment intends to manipulate cell suspensions extracted from whole blood to capture three-dimensional interactions of DNA within chromatin. This workflow has been written using NlaIII restriction enzyme and the heat denaturation method. For further information on protocol considerations, please see the Restriction Enzyme Pore-C info sheet.
Steps in the sequencing workflow
Prepare for your experiment You will need to:
- Prepare your samples for all experiments.
- Ensure you have your sequencing kits, the correct equipment and third-party reagents.
- Download the software for acquiring and analysing your data.
- Check your flow cells to ensure they have enough pores for a good sequencing run.
Library preparation
Duplex experiment: You will need to:
- Repair the DNA, and prepare the DNA ends for adapter attachment
- Attach sequencing adapters to the DNA ends
- Prime the flow cell, and load your DNA library into the flow cell
- Wash and reload the flow cell approximately every 20-24 hours
__Ultra-long DNA experiment:__ You will need to:
- Extract your uHMW gDNA
- Tagment your DNA using a diluted fragmentation mix
- Attach the sequencing adapters to the DNA ends
- Clean-up the sample by precipitating your DNA and elute overnight
- Prime the flow cell and load your DNA library into the flow cell
- Wash and reload the flow cell approximately every 20-24 hours
__Pore-C experiment:__ You will need to:
- Crosslink the three dimensional DNA interactions within the nucleus
- Permeabilise the cells to expose the crosslinked structures and denature the chromatin
- Cleave the genome into clusters of crosslinked DNA fragments at recognition sites with a restriction enzyme
- Ligate the cohesive ends of proximal crosslinked monomers into chimeric Pore-C polymers held in proximity
- Degrade the protein structures to release the chimeric Pore-C polymers into solution
- Purify the Pore-C extract before DNA repair and end-prep for adapter attachment
- Attach sequencing adapters to the DNA ends
- Prime the flow cell, and load your DNA library into the flow cell
- Wash and reload the flow cell approximately every 18 hours
Sequencing and analysis
You will need to:
- For each experiment, start a sequencing run using the MinKNOW software which will collect raw data from the device and convert it into basecalled reads.
- For each experiment, we recommend basecalling in real-time during sequencing before rebasecalling and aligning to a reference genome post-sequencing. Further details for each experiment set up are outlined in the "Sequencing and data analysis" section.
Compatibility of this protocol
This protocol should only be used in combination with:
- Ligation Sequencing Kit XL V14 (SQK-LSK114-XL)
- Ultra-Long DNA Sequencing Kit V14 (SQK-ULK114)
- Flow Cell Wash Kit XL (EXP-WSH004-XL)
- Flow Cell Priming Kit V14 (EXP-FLP004)
- Sequencing Auxiliary Vials V14 (EXP-AUX003)
- R10.4.1 PromethION Flow Cells (FLO-PRO114M)
2. Equipment and consumables
材料
- >15 ml of whole blood
- Ligation Sequencing Kit XL V14 (SQK-LSK114-XL)
- DNA 超长片段测序试剂盒(SQK-ULK114)
- Flow Cell Wash Kit XL (EXP-WSH004-XL)
耗材
- PromethION 测序芯片(FLO-PRO114M)
- Qubit dsDNA BR Assay(双链DNA宽范围检测)试剂盒(ThermoFisher ,Q32850)
- Qubit dsDNA HS Assay(双链DNA高灵敏度检测)试剂盒(Invitrogen, Q32851)
- Monarch® HMW DNA Extraction Kit for Tissue (NEB, T3060)
- Puregene Blood Kit (QIAGEN, 158023)
- T4 DNA Ligase 400,000 U/ml (NEB, M0202S/L)
- 供Oxford Nanopore Technologies®连接测序使用的NEBNext®配套模块(目录号E7180S或 E7180L),或使用以下三种NEBNext®产品:
- NEBNext® Ultra II 末端修复/ dA尾添加模块(NEB,E7546)
- NEBNext 快速连接模块(NEB,E6056)
- NEBNext FFPE DNA 修复混合液(NEB,M6630)
- 红细胞裂解液(QIAGEN,158904)
- Agencourt AMPure XP beads (Beckman Coulter, A63881)
- 5 M NaCl (Sigma, 71386)
- PEG 8000, 50% w/v (Rigaku Reagents, 25322-68-3)
- TE 缓冲液(10 mM Tris-HCl、1 mM EDTA、pH 8.0)(Fisher scientific,10224683)
- 0.5 M EDTA,pH 8(Thermo Scientific, R1021)
- Percoll, 1.135 g/ml (Cytiva, 17-0891-01)
- (Optional) dimethyl sulfoxide (DMSO) (Sigma-Aldrich, 20-139)
- ECOSURF EH-9 (Dow, 64366-70-7)
- Fetal bovine serum (FBS) (Gibco™, A3840401)
- Glycine (Sigma, 56-40-6)
- Formaldehyde at 36.5% v/v (Sigma, 33220)
- NlaIII restriction enzyme with CutSmart Buffer (NEB, R0125L)
- IGEPAL CA-630 (Sigma, I8896)
- Protease Inhibitor Cocktail (Sigma, P8340)
- Sodium dodecyl sulfate (SDS) at 10% v/v (Sigma, 71736)
- Tween-20 (Thermo Scientific, J20605.AP)
- 1 M Tris-HCl pH 8.0(Thermo Scientific, 15893661)
- Proteinase K at 20 μg/μl (NEB, P8107S)
- 10X phosphate-buffered saline (PBS), pH 7.4 (Thermo Fisher, 70011044)
- Recombinant Albumin at 20 μg/μl (NEB, B9200S)
- 无核酸酶水(如ThermoFisher,AM9937)
- 乙醇,100%(例如 Fisher, 16606002)
- 异丙醇,100%(Fisher, 10723124)
- Chilled phenol:chloroform:isoamyl alcohol in a 25:24:1 ratio, saturated with 10 mM Tris.HCl pH 8.0, 1 mM EDTA (Sigma, P3803-400ML)
- 3 M sodium acetate, pH 5.5 (Invitrogen, AM9740)
- 50 ml centrifuge tubes
- 15 ml Falcon离心管
- 2 ml Eppendorf DNA LoBind 离心管
- 0.2 ml 薄壁PCR管
- Qubit™ 分析管(Invitrogen, Q32856)
- Ziplock bags
- 0.2 µm filter
仪器
- PromethION 测序设备
- P1000 移液枪和枪头
- P200 移液枪和枪头
- P100 移液枪和枪头
- P10 移液枪和枪头
- P20 移液枪和枪头
- 宽口移液枪头
- Pasteur pipettes
- 热循环仪或恒温加热仪
- Hula混匀仪(低速旋转式混匀仪)
- 磁力架
- 涡旋混匀仪
- Temperature-controlled centrifuge
- 迷你离心机
- 盛有冰的冰桶
- Thermomixer
- Qubit™ 荧光计(或用于质控检测的等效仪器)
- Class I hood with active charcoal filter
- -80°C freezer storage
可选仪器
- Liquid nitrogen and canister
Approximately 15 ml of whole blood is required as input for the sample preparation steps.
Aliquots of whole blood are prepared for each experiment to isolate the peripheral blood mononuclear cells (PBMCs) for the Pore-C and Ultra-long DNA experiments and to extract gDNA for the Duplex experiment.
The whole blood can be collected in an anticoagulant such as K2-EDTA but we do not recommend mixing with other additives as they may interfere with the Pore-C DNA extraction or the DNA sequencing run.
We recommend different sample preparations due to different input requirements for each experiment. However, users may use other methods they feel are most appropriate but to ensure the individual input requirements are followed. Depending on how DNA is extracted from a sample, certain chemical contaminants may remain in the purified DNA, which can affect library preparation efficiency and sequencing quality. Read more about contaminants on the Contaminants page.
Input requirements:
- Pore-C experiment: 10 million PBMCs
- Duplex experiment: 1 μg or 100-200 fmol extracted DNA
- Ultra-long DNA experiment: 6 million PBMCs
We recommend preparing your samples and the custom SPRI bead suspension a day ahead of the experiments to ensure maximum use of time each day.
Third-party reagents
We have validated and recommend the use of all the third-party reagents used in this protocol. Alternatives have not been tested by Oxford Nanopore Technologies.
For all third-party reagents, we recommend following the manufacturer's instructions to prepare the reagents for use.
NEBNext® Companion Module for Oxford Nanopore Technologies® Ligation Sequencing
We recommend buying the NEBNext® Companion Module for Oxford Nanopore Technologies® Ligation Sequencing (catalogue number E7180S or E7180L). This contains all the NEB reagents needed for use with the Ligation Sequencing Kit XL V14 (SQK-LSK114-XL) during day 1 of the Duplex experiment and day 4 of the Pore-C experiment.
Please note, the Pore-C experiment also requires additional use of the T4 DNA Ligase kit for the proximity ligase reaction during day 2 of the protocol.
Ligation Sequencing Kit XL V14 (SQK-LSK114-XL) contents
| Name | Acronym | Vial colour | Number of vials | Fill volume per vial (µl) |
|---|---|---|---|---|
| DNA Control Strand | DCS | Yellow | 1 | 100 |
| Ligation Adapter | LA | Green | 1 | 320 |
| Ligation Buffer | LNB | White | 1 | 1,500 |
| Elution Buffer | EB | White cap, black strip label | 1 | 10,000 |
| Long Fragment Buffer | LFB | White cap, orange strip label | 2 | 20,000 |
| Short Fragment Buffer | SFB | White cap, blue strip label | 2 | 20,000 |
| Library Beads | LIB | Pink | 2 | 1,800 |
| Library Solution | LIS | White cap, pink label | 2 | 1,800 |
| Sequencing Buffer | SB | Red | 3 | 1,700 |
| Flow Cell Flush | FCF | Clear | 4 | 15,500 |
| Flow Cell Tether | FCT | Purple | 1 | 1,600 |
Note: The DNA Control Sample (DCS) is a 3.6 kb standard amplicon mapping the 3' end of the Lambda genome.
Ultra-Long DNA Sequencing Kit (SQK-ULK114) contents
We had previously uncovered an issue with the Precipitation Stars (PS) found in the SQK-ULK114 kits and removed their use from the method to prevent any potential issues.
We have since improved our manufacturing and internal validation processes of the Precipitation Stars (PS) and are now in the position to reintroduce their use with the SQK-ULK114 Kits.
Kit format with improved precipitation stars that can be used:
Batch ULK114.30.0001 or newer

| Name | Acronym | Cap colour | Number of vials | Fill volume per vial (µl) |
|---|---|---|---|---|
| Rapid Adapter | RA | Green | 1 | 40 |
| Fragmentation Mix | FRA | Amber | 1 | 50 |
| FRA Dilution Buffer | FDB | Clear | 1 | 1,600 |
| Elution Buffer | EB | Black | 2 | 1,500 |
| Extraction EB | EEB | Orange | 3 | 1,700 |
| Sequencing Buffer UL | SBU | Red | 2 | 1,000 |
| Loading Solution UL | LSU | White cap, pink label | 1 | 200 |
| Flush Tether UL | FTU | Purple | 1 | 600 |
| Flow Cell Flush | FCF | Blue | 2 | 15,500 |
| Precipitation Buffer | PTB | Blue | 2 | 1,700 |
| Precipitation Star | PS | Yellow | 6 | 1 star |
Kit format where stars should not be used:
Batch ULK114.20.xxxx or older:
| Name | Acronym | Cap colour | Number of vials | Fill volume per vial (µl) |
|---|---|---|---|---|
| Rapid Adapter | RA | Green | 1 | 40 |
| Fragmentation Mix | FRA | Amber | 1 | 50 |
| FRA Dilution Buffer | FDB | Clear | 1 | 1,600 |
| Elution Buffer | EB | Black | 2 | 1,500 |
| Extraction EB | EEB | Orange | 3 | 1,700 |
| Sequencing Buffer UL | SBU | Red | 2 | 1,000 |
| Loading Solution UL | LSU | White cap, pink label | 1 | 200 |
| Flush Tether UL | FTU | Purple | 1 | 600 |
| Flow Cell Flush | FCF | Blue | 2 | 15,500 |
| Precipitation Buffer | PTB | Blue | 2 | 1,700 |
| Precipitation Star | PS | Yellow | 6 | 1 star |
Flow Cell Wash Kit XL (EXP-WSH004-XL) contents
- Wash Mix (WMX) contains DNase I.
- Wash Diluent (DIL) contains the exonuclease buffer that maximises activity of the DNase I.
- The Storage Buffer allows flow cells to be stored for extended periods of time.
3. Computer requirements and software
PromethION 24/48 IT requirements
The PromethION device contains all the hardware required to control up to 24 (for the P24 model) or 48 (for the P48 model) sequencing experiments and acquire the data. The device is further enhanced with high performance GPU technology for real-time basecalling. Read more in the PromethION IT requirements document.
PromethION 2 Solo IT requirements
The PromethION 2 (P2) Solo is a device which directly connects into a GridION Mk1 or a stand-alone computer that meets the miminum specifications for real-time data streaming and analysis. Up to two PromethION flow cells can be can be run and each is independently addressable, meaning experiments can be run concurrently or individually. For information on the computer IT requirements, please see the PromethION 2 Solo IT requirements document.
Software for nanopore sequencing
MinKNOW
The MinKNOW software controls the nanopore sequencing device, collects sequencing data and basecalls in real time. You will be using MinKNOW for every sequencing experiment to sequence, basecall and demultiplex if your samples were barcoded.
For instructions on how to run the MinKNOW software, please refer to the MinKNOW protocol.
EPI2ME (optional)
The EPI2ME cloud-based platform performs further analysis of basecalled data, for example alignment to the Lambda genome, barcoding, or taxonomic classification. You will use the EPI2ME platform only if you would like further analysis of your data post-basecalling.
For instructions on how to create an EPI2ME account and install the EPI2ME Desktop Agent, please refer to this link.
Check your PromethION flow cell
We highly recommend that you check the number of pores in your flow cell prior to starting a sequencing experiment. This should be done within 12 weeks of purchasing for PromethION flow cells. Oxford Nanopore Technologies will replace any flow cell with fewer than 5000 pores when the result is reported within two days of performing the flow cell check, and when the storage recommendations have been followed. To do the flow cell check, please follow the instructions in the Flow Cell Check document.
4. Custom SPRI bead preparation for the Pore-C experiment
耗材
- Agencourt AMPure XP beads (Beckman Coulter, A63881)
- 1 M Tris-HCl, pH 7.5
- 0.5 M EDTA,pH 8(Thermo Scientific, R1021)
- 5 M NaCl (Sigma, 71386)
- PEG 8000, 50% w/v (Rigaku Reagents, 25322-68-3)
- 无核酸酶水(如ThermoFisher,AM9937)
- 新制备的 80% 乙醇(用无核酸酶水配制)
- 2 ml Eppendorf DNA LoBind 离心管
仪器
- 磁力架
- Hula混匀仪(低速旋转式混匀仪)
- 热循环仪或恒温加热仪
- P1000 移液枪和枪头
- P200 移液枪和枪头
- P100 移液枪和枪头
- P10 移液枪和枪头
- 宽口移液枪头
Custom SPRI bead suspension preparation for Pore-C extraction
Before starting the Pore-C experiment, a custom SPRI bead suspension needs to be prepared. This will be used to deplete non-chimeric monomers and to maximise the frequency of chimeric Pore-C polymers, improving purity ratios and read lengths at the end of Day 3: Pore-C experiment.
We also recommend using this custom SPRI bead suspension to size-select the extracted gDNA input in the Whole blood sample gDNA extraction for the Duplex experiment to enrich for fragments above 1.5-2 kb.
Store the beads at 4°C and bring to room temperature before use.
Prepare a custom buffer in a 2 ml Eppendorf DNA LoBind tube as follows for use in step 7.
| Reagent | Final | Volume |
|---|---|---|
| Tris-HCl, 1 M | 10 mM | 20 μl |
| EDTA, pH 8, 0.5 M | 1 mM | 4 μl |
| NaCl, 5 M | 1.6 M | 640 μl |
| PEG 8000, 50% (w/v) | 11% (w/v) | 440 μl |
| Nuclease-free water | - | 888 μl |
| Total | - | 1992 μl |
Note: We recommend using wide-bore 1 ml pipette tips to accurately pipette 440 μl of 50% PEG 8000.
Transfer 1 ml of resuspended Agencourt AMPure XP beads into two 2 ml Eppendorf DNA LoBind tubes, so that each tube contains 1 ml.
Place the tubes on a magnetic rack to pellet the beads until the solution is clear and colourless. Pipette off and discard the supernatant.
Remove the tubes from the magnet and resuspend the pellets with 1 ml of nuclease-free water. Pellet the beads on the magnet until supernatant is clear and colourless and pipette off the supernatant.
Repeat the previous step.
Spin down and place the tubes back on the magnet to pipette off any residual water.
Resuspend both tubes of pelleted beads in 200 µl of custom buffer and then pool both tubes into a single tube to a total of 400 µl.
Transfer the remaining custom buffer into the tube containing the pooled beads.
Store the beads at 4°C. Before use, bring the suspension to room temperature.
5. Whole blood sample preparation for the Pore-C experiment
材料
- 5–10 ml whole blood
耗材
- 10X phosphate-buffered saline (PBS), pH 7.4 (Thermo Fisher, 70011044)
- Percoll, 1.135 g/ml (Cytiva, 17-0891-01)
- (Optional) dimethyl sulfoxide (DMSO) (Sigma-Aldrich, 20-139)
- Fetal bovine serum (FBS) (Gibco™, A3840401)
- (Optional) chilled fetal bovine serum (FBS) (Gibco™, A3840401)
- 50 ml centrifuge tubes
- 2 ml Eppendorf DNA LoBind 离心管
仪器
- Pasteur pipettes
- Temperature-controlled centrifuge with rotor for 2 ml and 50 ml tubes
- 盛有冰的冰桶
- P1000 移液枪和枪头
- P200 移液枪和枪头
- P100 移液枪和枪头
- P20 移液枪和枪头
- P10 移液枪和枪头
可选仪器
- Liquid nitrogen and canister
- -80°C freezer storage
PBMC sample preparation for Pore-C DNA extraction
Before starting the Pore-C DNA extraction, the peripheral blood mononuclear cells (PBMCs) must be isolated from whole blood whilst maintaining cell viability. Approximately 5–10 ml of whole blood should yield sufficient PBMCs for the Pore-C DNA extraction. If necessary, combine multiple aliquots of whole blood to achieve a final 5–10 ml pooled sample. Using the below method, approximately 10 million PBMCs are prepared in aliquots of 1X PBMCs supplemented with 2% FBS. Approximately 10 million PBMCs are taken forwards into the day 1 of the Pore-C experiment.
Users may isolate PBMCs by any means they feel are most appropriate for the whole blood sample to be used, provided that:
- PBMCs are isolated as soon as possible from fresh blood and no later than 24 hours.
- PBMC are isolated using a method optimised for cell viability.
- The whole blood is not mixed with any additives, except for anticoagulants (e.g. K2-EDTA), which are acceptable and will not interfere with the Pore-C DNA extraction.
If users have a minimum of approximately 10 million PBMCs which have been isolated previously which satisfy these requirements, they may start sample extraction directly from day 1 of the Pore-C experiment.
Prepare three solutions in preparation for white blood cells isolation:
- 500 ml of 1X PBS supplemented with 2% FBS final concentration and store at room temperature.
| Reagent | Volume |
|---|---|
| 10X PBS | 50 ml |
| Fetal bovine serum (FBS) | 10 ml |
| Nuclease-free water | 440 ml |
| Total | 500 ml |
- 100 ml of 1X PBS supplemented with 60% Percoll final concentration and store at room temperature.
| Reagent | Volume |
|---|---|
| 10X PBS | 10 ml |
| Percoll | 60 ml |
| Nuclease-free water | 30 ml |
| Total | 100 ml |
- (Optional for storage) 2 ml of FBS supplemented with 20% DMSO final and store at 4°C.
| Reagent | Volume |
|---|---|
| FBS | 1,600 µl |
| DMSO | 400 µl |
| Total | 2,000 µl |
Allow the whole blood sample to warm to room temperature and then dilute with equal volume of room temperature 1X PBS supplemented with 2% FBS. Transfer the diluted blood to a 50 ml centrifuge tube.
Centrifuge at 800 x g at 20°C for 10 minutes with the brake off to prevent remixing of the separated fractions.
After centrifugation, the whole blood should have separated into the plasma, buffy coat and red blood cells. Check the turbidity of the plasma layer (the top layer). If it is not clear, centrifuge at 800 x g at 20°C for a further 10 minutes with the brake off.

Using a Pasteur pipette, remove as much of the plasma layer as possible without disturbing the layer of buffy coat. Gently remove the buffy coat layer, taking care to draw as little of the red blood cell layer as possible. Transfer the recovered buffy coat to a fresh 50 ml centrifuge tube.
Make up the recovered buffy coat sample to 25 ml of 1X PBS supplemented with 2% FBS.
Aliquot 20 ml of 1X PBS supplemented with 60% Percoll in a fresh 50 ml centrifuge tube.
Using a fresh Pasteur pipette, very gently layer the diluted buffy coat sample over the Percoll layer at a 45° angle.

Centrifuge at 350 x g at 20°C for 40 minutes with slow acceleration and with the brake off.
Check the turbidity of the plasma layer and the formation of the white blood cells layer. If the plasma layer is not clear or the PBMC layer is not well defined, continue to centrifuge at 350 x g at 20°C for a further 20 minutes using slow acceleration with the brake off.

Using a Pasteur pipette, remove as much of the plasma layer as possible without disturbing the layer of white blood cells, then gently remove the layer of PBMCs. It is acceptable to draw plasma with the layer of white blood cells; however, take care to draw as little of the Percoll layer as possible.
Transfer the recovered white blood cells to a fresh 50 ml centrifuge tube.
Resuspend the recovered white blood cells in 50 ml of room temperature 1X PBS supplemented with 2% FBS.
Centrifuge at 350 x g at 20°C for 15 minutes with the brake on.
Aspirate and discard the supernatant. Gently resuspend the white blood cells in 25 ml of room temperature 1X PBS supplemented with 2% FBS. Centrifuge at 350 x g at 20°C for 15 minutes with the brake on.
Repeat the previous step.
Aspirate and discard the supernatant. Gently resuspend the white blood cells in another 25 ml of room temperature 1X PBS supplemented with 2% FBS.
Centrifuge at 200 x g at 20°C for 10 minutes with the brake on.
Assuming every 1 ml of whole blood originally used will yield approximately 1.5 million white blood cells, resuspend cells to approximately 10 million white blood cells/ml in room temperature 1X PBS supplemented with 2% FBS.
Transfer an aliquot of approximately 10 million white blood cells total to a fresh 2 ml Eppendorf DNA LoBind tube.
Cool on ice for 5 minutes.
The cells can be stored if Pore-C sample extraction cannot be started immediately.
- Centrifuge at 350 x g at 4°C for 2 minutes with the brake on.
- Aspirate and discard the supernatant, then resuspend the white blood cells pellet in 1 ml of chilled FBS.
- Once resuspended, slowly mix in 1 ml of chilled FBS supplemented with 20% DMSO, drop by drop.
Note: As DMSO is mixed with water, energy is released as heat. Adding DMSO to the white blood cells suspension drop by drop prevents heat shock to the cells. - Snap freeze aliquots of white blood cells in liquid nitrogen then store at –80°C.
Take forward approximately 10 million white blood cells into the Pore-C experiment. Store the cells at 4°C until the experiment can begin.
6. Whole blood sample gDNA extraction for the Duplex experiment
材料
- 3 ml of whole blood
- Custom SPRI bead suspension
耗材
- Puregene Blood Kit (QIAGEN, 158023)
- 新制备的 80% 乙醇(用无核酸酶水配制)
- 异丙醇,100%(Fisher, 10723124)
- TE 缓冲液(10 mM Tris-HCl、1 mM EDTA、pH 8.0)(Fisher scientific,10224683)
- 1.5 ml Eppendorf DNA LoBind 离心管
- 15 ml Falcon离心管
- Qubit™ 分析管(Invitrogen, Q32856)
- Qubit dsDNA HS Assay(双链DNA高灵敏度检测)试剂盒(Invitrogen, Q32851)
仪器
- 适用于 15 ml Falcon 管的离心机及转子
- Heating block
- 涡旋混匀仪
- Qubit™ 荧光计(或用于质控检测的等效仪器)
Whole blood gDNA extraction for the Duplex experiment
An input of 1 µg of gDNA must be prepared for the Duplex experiment. You may extract the gDNA by any means is most appropriate. This section outlines how to extract gDNA from whole blood using the QIAGEN Puregene Blood Kit.
Perform cell separation and lysis according to the QIAGEN Puregene Handbook for 3 ml of blood (pages 19–20, steps 1–7):
Dispense 9 ml of RBC Lysis Solution into a 15 ml Falcon tube.
Add 3 ml of whole blood and mix by inverting 10 times.
Incubate for 5 minutes at room temperature. Invert at least once during the incubation.
Centrifuge for 2 minutes at 2000 x g to pellet the white blood cells.
Carefully discard the supernatant by pipetting or pouring, leaving approximately 200 µl of the residual liquid and the white blood cell pellet.
Vortex the tube vigorously to resuspend the pellet in the residual liquid. Vortexing greatly facilitates cell lysis in the next step.
Add 3 ml of Cell Lysis Solution and pipette mix to lyse the cells or vortex for 10 seconds.
Incubate the samples at 37°C for 30 minutes. If the sample is not homogenous, gently invert the tubes and extend the incubation for another 30 minutes.
Purify the lysate according to the QIAGEN Puregene Handbook for 3 ml of blood (pages 20–21, steps 8–17):
Add 15 µl of RNase A Solution and mix by inverting 25 times. Incubate for 15 minutes at 37°C. Then incubate for 3 minutes on ice to quickly cool the sample.
Add 1 ml of Protein Precipitation Solution and vortex vigorously for 20 seconds at high speed.
Centrifuge for 5 minutes at 2000 x g. The precipitated proteins should form a tight brown pellet. If the protein pellet is not tight, incubate on ice for 5 minutes and repeat the centrifugation.
Pipette 3 ml of isopropanol into a clean 15 ml Falcon tube and add the supernatant from the previous step by pouring carefully. Be sure that the protein pellet is not dislodged during pouring.
Mix by inverting 50 times until the DNA is visible as threads or a clump.
Centrifuge for 3 minutes at 2000 x g. The DNA may be visible as a small white pellet.
Carefully discard the supernatant and drain the tube by inverting on a clean piece of absorbent paper, taking care that the pellet remains in the tube.
Add 3 ml of 80% ethanol and invert several times to wash the DNA pellet.
Centrifuge for 1 minute at 2000 x g.
Carefully discard the supernatant. Drain the tube on a clean piece of absorbent paper, taking care that the pellet remains in the tube. Dry the pellet for 5-10 minutes. The pellet might be loose and easily dislodged. Avoid over-drying the DNA pellet as the DNA will be difficult to dissolve.
To maximise the DNA yield, we recommend that the elution is performed for 2 hours at 50°C, using 150 µl TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0), occasionally mixing the tube contents by gentle inversion.
Quantify 1 µl of eluted sample using a Qubit fluorometer.
Dilute your DNA sample to 60 ng/µl in a final volume of 50 µl of TE buffer at pH 8.
Add 35 µl of room temperature custom SPRI bead suspension to your DNA sample, and mix by flicking the tube.
Incubate on a Hula mixer (rotator mixer) for 10 minutes at room temperature.
Spin down briefly and pellet on a magnet until the supernatant is clear and colourless. Keep the tube on the magnet, and pipette off the supernatant.
Keep the tube on the magnet and wash the beads with 200 µl of freshly prepared 80% ethanol without disturbing the pellet. Remove the ethanol using a pipette and discard.
Repeat the previous step.
Spin down and place the tube back on the magnetic rack. Pipette off any residual ethanol. Allow the pellet to dry for ~30 seconds, but do not dry the pellet to the point of cracking.
Remove the tube from the magnetic rack and resuspend the pellet in 40 µl of TE buffer. Incubate for 1 minute at 50°C, and then for 5 minutes at room temperature.
Pellet the beads on a magnet until the eluate is clear and colourless, for at least 1 minute.
Remove and retain 40 µl of eluate into a clean 1.5 ml Eppendorf DNA LoBind tube.
Quantify 1 µl of size-selected DNA using a Qubit fluorometer.
You can expect a 50-55% loss of DNA depending on a fragment length distribution of input material: the greater the proportion of short fragments (<1.5-2 kb), the greater the sample loss.
Take forward 1 µg of extracted DNA into the Duplex experiment. Store at 4°C until the experiment can begin.
7. Whole blood sample preparation for the Ultra-long DNA experiment
材料
- 1.6 ml 全血
耗材
- 红细胞裂解液(QIAGEN,158904)
- 10X phosphate-buffered saline (PBS), pH 7.4 (Thermo Fisher, 70011044)
- 15 ml Falcon离心管
- 1.5 ml Eppendorf DNA LoBind 离心管
仪器
- Temperature-controlled microfuge
- P1000 移液枪和枪头
- P200 移液枪和枪头
- P20 移液枪和枪头
PBMC sample preparation for the Ultra-long DNA experiment
Approximately 6 million isolated PBMCs must be prepared from 1.6 ml of whole blood to use as input in the Ultra-long DNA experiment.
Users may isolate PBMCs by any means they feel are most appropriate for the whole blood sample to be used. If 6 million cells have been isolated, users can start from day 1 of the Ultra-long DNA experiment.
In a fresh 15 ml Falcon tube, prepare 10 ml of 1x PBS in nuclease-free water as follows:
| Reagent | Volume |
|---|---|
| 10X PBS | 1 ml |
| Nuclease-free water | 9 ml |
| Total | 10 ml |
Add 4.8 ml of RBC Lysis Solution to 1.6 ml of whole blood in a 15 ml Falcon tube.
Gently invert the tube ten times to mix.
Incubate for 5 minutes at room temperature and gently invert twice during the incubation.
Centrifuge at 2000 x g for 2 minutes at 4°C to pellet the white blood cells.
Discard the supernatant by pouring. There will be ~200 µl supernatant remaining in the tube.
Resuspend the cells in the residual supernatant by gently flicking the tube.
Make up the volume to 1.6 ml with 1x PBS.
Repeat steps 1-7 twice more to complete three washes in total.
If any red colouration persists, repeat the wash step until the cell pellet is white.
After the final spin, remove the entire supernatant by pouring and aspirating any remaining supernatant.
Resuspend the cell pellet in 40 µl 1x PBS. There will be approximately 6 million cells in the suspension.
Take forward 6 million PBMCs forward into the Ultra-Long DNA experiment. Store the pellet at 4°C until the experiment can begin.
8. Day 1: Pore-C experiment
材料
- 10 million white blood cells isolated from whole blood
耗材
- 无核酸酶水(如ThermoFisher,AM9937)
- NlaIII restriction enzyme with CutSmart Buffer (NEB, R0125L)
- ECOSURF EH-9 (Dow, 64366-70-7)
- Glycine (Sigma, 56-40-6)
- Formaldehyde at 36.5% v/v (Sigma, 33220)
- IGEPAL CA-630 (Sigma, I8896)
- Protease Inhibitor Cocktail (Sigma, P8340)
- Chilled 10X phosphate-buffered saline (PBS) (Thermo Fisher, 70011044)
- Sodium dodecyl sulfate (SDS) at 10% v/v (Sigma, 71736)
- 5 M NaCl (Sigma, 71386)
- 1 M Tris-HCl pH 8.0(Thermo Scientific, 15893661)
- Ziplock bags
- 0.2 µm filter
- 50 ml centrifuge tubes
- 2 ml Eppendorf DNA LoBind 离心管
- 1.5 ml Eppendorf DNA LoBind 离心管
仪器
- Class I hood with active charcoal filter
- Temperature-controlled centrifuge
- 热循环仪或恒温加热仪
- Thermomixer
- 涡旋混匀仪
- Hula混匀仪(低速旋转式混匀仪)
- 盛有冰的冰桶
- P1000 移液枪和枪头
- P100 移液枪和枪头
- P200 移液枪和枪头
- P20 移液枪和枪头
- 宽口移液枪头
Day 1: Pore-C experiment overview
During day 1, the PBMCs are prepared for stabilising of the three-dimensional interactions of DNA in the nucleus by chemically cross-linking DNA and protein. The nuclei are then permeabilised to expose the crosslinked cytoskeleton cage and nuclear structures before the chromatin is denatured. The DNA is now accessible to the chosen restriction enzyme† which passively diffuses through the crosslinked cytoskeleton cage and nuclear structures to digest the genome at compatible recognition sites. The sample is incubated overnight which creates clusters of DNA fragments held in proximity by crosslinks between DNA and the cytoskeleton, preserving the original interactions which were crosslinked.
†This protocol has been written using NlaIII and the heat denaturation method as our investigations have found this 4-cutter is particularly suitable for Pore-C across many different species, yielding Pore-C extracts with high contact densities. For more information, please see the "Protocol considerations" section of our Restriction enzyme Pore-C info sheet.
Thaw the NlaIII restriction enzyme and CutSmart Buffer in accordance with the manufacturer's instructions and place on ice.
- Thaw both reagents on ice.
- Flick and/or invert the reagent tubes to ensure they are well mixed.
Note: Do not vortex the NlaIII restriction enzyme. - Spin down tubes before opening for the first time each day.
Prepare 1 ml of 1% SDS in nuclease-free water, as follows:
| Reagent | Volume |
|---|---|
| 10% SDS | 100 µl |
| Nuclease-free water | 900 µl |
| Total | 1,000 µl |
Prepare 10 ml of 10% (v/v) ECOSURF™ EH-9 in nuclease-free water, as follows:
- Weigh out 1 g of ECOSURF™ EH-9.
- Transfer to a fresh 15 ml Falcon tube.
- Add 9 ml of nuclease-free water.
- Gently pipette mix with a wide-bore pipette tip until the solution is homogenous.
Prepare 1 ml of 2.5 M glycine filtered through a 0.2 µm filter and store at room temperature.
Prepare 200 ml filtered 1X PBS and chill at 4°C.
Pre-cool a centrifuge to 4°C.
1% formaldehyde solution is a biological hazard. Formaldehyde crosslinks DNA and is a mutagen. It must be handled with caution, and vessels containing the solution should only be uncapped in a class I hood.
Prepare the formaldehyde solution as follows:
Transfer 10 ml of 1X PBS into a 50 ml Falcon tube. Note: Using a 15 ml Falcon tube is not recommended.
Inside a class I hood, with double gloves, add 291 μl of 36.5% formaldehyde to the 10 ml 1X PBS aliquot to a final concentration of 1% formaldehyde in ~10.3 ml.
Mix by gentle inversion, and open the tube to allow gases to escape, then close the tube.
Check that no formaldehyde residue has remained on the gloves, Falcon tube, or pipette.
Remove the outer gloves and discard them in a biohazard bag in the hood.
Remove the 1% formaldehyde 1X PBS solution from the hood.
Store the tube with formaldehyde inside a zip lock bag at 4°C prior to use.
Prepare the white blood cells as follows:
Take approximately 10 million white blood cells and briefly homogenise the suspension by gently pipetting with a wide-bore pipette tip.
Transfer the cell suspension to a 50 ml centrifuge tube.
Rinse the original tube with a further 1 ml of chilled 1X PBS into the 50 ml centrifuge tube.
Bring the volume of the resuspended white blood cells to 10 ml in chilled 1X PBS.
Proceed with the Pore-C experiment using approximately 10 million white blood cells as input.
Centrifuge the sample at 300 x g at 4°C for 5 minutes.
Aspirate and discard the supernatant, then add 10 ml of chilled 1X PBS to the pellet. Resuspend the pellet by gently pipetting up and down using a wide-bore pipette tip.
Centrifuge the sample at 300 x g at 4°C for 5 minutes.
Check the 2.5 M glycine solution has not precipitated before crosslinking the sample. Dissolve precipitate with heat and vortexing if required.
Inside a class I hood, with double gloves, aspirate and discard the supernatant.
Add 1 ml of the previously prepared 1% formaldehyde solution 1X PBS to the pellet. Resuspend the pellet by gently pipetting up and down using a wide-bore pipette tip.
Once resuspended, add a further 9 ml of the 1% formaldehyde solution in 1X PBS. Mix gently by pipetting up and down, using a wide-bore pipette tip.
Incubate at room temperature for exactly 10 minutes to crosslink the sample. The incubated solution should be mixed by gentle agitation every few minutes.
We do not recommend extending incubation times as it may have a detrimental impact on the efficiency of de-crosslinking the DNA later in the protocol.
Inside the hood with double gloves, quench the formaldehyde by adding 527 μl of 2.5 M glycine to the sample suspension for a final concentration of 1% w/v glycine (125 mM) in ~10.5 ml. Mix gently by pipetting up and down, using a wide-bore pipette tip.
Incubate at room temperature for 5 minutes, then chill on ice for a further 10 minutes with regular, gentle agitation.
Centrifuge the crosslinked sample suspension at 300 x g at 4°C for 5 minutes.
Continuing in the class I hood, aspirate and discard the supernatant. Add 10 ml of chilled 1X PBS to the tube.
Centrifuge the sample at 500 x g at 4°C for 5 minutes.
Continuing in the class I hood, aspirate and discard the supernatant, and add 1 ml of chilled 1X PBS to the pellet. Mix gently by pipetting up and down using a wide-bore pipette tip.
Split the resuspended sample into two separate 500 μl aliquots in fresh 2 ml Eppendorf tubes.
Note: 2 ml Eppendorf tubes are required for a compact sample pellet. Do not use 1.5 ml tubes.
Wash the previous sample tube with a further 1 ml of 1X PBS, and split this between the two aliquots in 2 ml Eppendorf DNA LoBind tubes.
The rest of the protocol can be continued outside of the class I hood.
Centrifuge the samples at 500 x g at 4°C for 5 minutes. Aspirate and discard the supernatant.
Process each crosslinked sample pellet separately. Do not pool multiple pellets into a single reaction.
The remainder of the protocol is written for one pellet.
We advise continuing with a freshly crosslinked sample pellet. However, if you intend to store samples for later use, you can snap-freeze the aliquots in liquid nitrogen. Store frozen sample pellets at –80°C and use within one year.
Do not proceed any further unless it is possible to complete the remainder of this section consecutively without interruption. It is not advisable to incubate any step longer than stated in this protocol. Doing so may be detrimental to Pore-C data quality and sequencing performance.
Pre-cool a microfuge to 4°C and set a thermomixer to 65°C.
Prepare 600 μl of 1.5X CutSmart Buffer in nuclease-free water as follows in a 1.5 ml Eppendorf DNA LoBind tube. Keep on ice.
| Reagent | Volume |
|---|---|
| Nuclease-free water | 510 µl |
| 10X CutSmart Buffer | 90 µl |
| Total | 600 µl |
To make the permeabilisation solution, add the components below to a 1.5 ml Eppendorf DNA LoBind tube in the following order. Keep the prepared permeabilisation solution on ice at 4°C until ready to use.
| Reagent | Final | Volume |
|---|---|---|
| Tris-HCl, pH 8.0, 1 M | 10 mM | 5 µl |
| NaCl, 5 M | 10 mM | 1 µl |
| IGEPAL CA-630, 10% | 0.2% | 10 µl |
| Nuclease-free water | - | 484 µl |
| Total | - | 500 µl |
Thaw the protease inhibitor cocktail on ice and spin down.
Add 50 μl of protease inhibitor cocktail to 500 μl of permeabilisation solution at 4°C.
Add 550 μl protease inhibitor cocktail-permeabilisation solution to the sample pellet. Resuspend the pellet by gently pipetting up and down, using a wide-bore pipette tip.
Incubate on ice for 15 minutes and mix by regular, gentle inversion.
Centrifuge the sample at 500 x g at 4°C for 10 minutes.
Following centrifugation, the pellet will be delicate. Carefully aspirate and discard as much of the supernatant as possible without disturbing the pellet
Resuspend the pellet in 200 μl of the prepared chilled 1.5X CutSmart buffer by gently pipetting up and down, using a wide-bore pipette tip.
Centrifuge the sample at 500 x g at 4°C for 5 minutes. Aspirate and discard the supernatant.
Resuspend the pellet in 300 μl of the prepared chilled 1.5X CutSmart buffer by gently pipetting up and down, using a wide-bore pipette tip.
To denature the chromatin, add 33.5 μl 1% SDS directly to the sample suspension to a final concentration of 0.1% SDS and a total volume of 333.5 μl. Mix gently by pipetting up and down using a wide-bore pipette tip.
The SDS may precipitate at this point; this will not impact the experiment so proceed to the next step.
Incubate the sample suspension in a thermomixer at 300 RPM at 65°C for 10 minutes.
Note: This incubation can be performed without mixing.
Remove the tube from the thermomixer and immediately put on ice.
Set the thermomixer to 37°C.
Add 37.5 μl of 10% (v/v) ECOSURF™ EH-9 directly to the cell suspension for a final concentration of 1% ECOSURF™ EH-9 (total volume of 371 μl). Mix gently by pipetting with a wide-bore pipette tip.
Incubate the tube on ice for 10 minutes.
The SDS may precipitate at this point. This will not impact the experiment so proceed to the next step.
Add the following reagents to the sample suspension and invert 3-4 times to mix.
| Reagent | Final | Volume |
|---|---|---|
| Permeabilised cells | - | 371 µl |
| NEB NlaIII, 10 U/µl | 1 U/µl | 45 µl |
| Nuclease-free water | - | 34 µl |
| Total | - | 450 µl |
Incubate the tube in a thermomixer at 37°C for 18 hours with periodic <1000 rpm rotation for <30 seconds every 15 minutes. This will prevent condensation inside the lid.
Note: This incubation can be performed without mixing.
During the Pore-C incubation, start the Duplex experiment.
9. Day 1: Duplex experiment
材料
- 1 μg or 100-200 fmol DNA
- Ligation Sequencing Kit XL V14 (SQK-LSK114-XL)
耗材
- NEBNext FFPE DNA 修复混合液(NEB,M6630)
- NEBNext® Ultra II 末端修复/ dA尾添加模块(NEB,E7546)
- NEBNext 快速连接模块(NEB,E6056)
- Qubit dsDNA HS Assay(双链DNA高灵敏度检测)试剂盒(Invitrogen, Q32851)
- Agencourt AMPure XP 磁珠(Beckman Coulter™, A63881)
- 无核酸酶水(如ThermoFisher,AM9937)
- 新制备的 80% 乙醇(用无核酸酶水配制)
- Qubit™ 分析管(Invitrogen, Q32856)
- 1.5 ml Eppendorf DNA LoBind 离心管
- 0.2 ml 薄壁PCR管
仪器
- P1000 移液枪和枪头
- P100 移液枪和枪头
- P20 移液枪和枪头
- P10 移液枪和枪头
- 迷你离心机
- Hula混匀仪(低速旋转式混匀仪)
- 磁力架
- 盛有冰的冰桶
- 涡旋混匀仪
- 热循环仪
- Qubit™ 荧光计(或用于质控检测的等效仪器)
Day 1: Duplex experiment overview
In this step, the library for the Duplex experiment is prepared for sequencing, as follows: the extracted DNA is repaired and the ends prepared for adapter attachment using the NEBNext FFPE DNA Repair Mix and NEBNext Ultra II End Repair/dA-tailing Module reagents. The sequencing adapters are attached to the DNA fragment ends before a clean-up step in preparation for sequencing on the PromethION Flow Cell.
Thaw DNA Control Sample (DCS) at room temperature, spin down, mix by pipetting, and place on ice.
We recommend using the DNA Control Sample (DCS) in your library prep for troubleshooting purposes. However, you can omit this step and make up the extra 1 µl with your sample DNA.
Prepare the NEBNext FFPE DNA Repair Mix and NEBNext Ultra II End Repair / dA-tailing Module reagents in accordance with manufacturer’s instructions, and place on ice.
For optimal performance, NEB recommend the following:
Thaw all reagents on ice.
Flick and/or invert the reagent tubes to ensure they are well mixed.
Note: Do not vortex the FFPE DNA Repair Mix or Ultra II End Prep Enzyme Mix.Always spin down tubes before opening for the first time each day.
The Ultra II End Prep Buffer and FFPE DNA Repair Buffer may have a little precipitate. Allow the mixture to come to room temperature and pipette the buffer up and down several times to break up the precipitate, followed by vortexing the tube for 30 seconds to solubilise any precipitate.
Note: It is important the buffers are mixed well by vortexing.The FFPE DNA Repair Buffer may have a yellow tinge and is fine to use if yellow.
Prepare the DNA in nuclease-free water:
- Transfer 1 μg (or 100-200 fmol) input DNA into a 1.5 ml Eppendorf DNA LoBind tube.
- Adjust the volume to 47 μl with nuclease-free water.
- Mix thoroughly by pipetting up and down, or by flicking the tube.
- Spin down briefly in a microfuge
In a 0.2 ml thin-walled PCR tube, mix the following:
Between each addition, pipette mix 10-20 times.
| Reagent | Volume |
|---|---|
| DNA from the previous step | 47 µl |
| DNA CS (optional) | 1 µl |
| NEBNext FFPE DNA Repair Buffer | 3.5 µl |
| NEBNext FFPE DNA Repair Mix | 2 µl |
| Ultra II End-prep Reaction Buffer | 3.5 µl |
| Ultra II End-prep Enzyme Mix | 3 µl |
| Total | 60 µl |
Thoroughly mix the reaction by gently pipetting and briefly spinning down.
Using a thermal cycler, incubate at 20°C for 5 minutes and 65°C for 5 minutes.
Resuspend the AMPure XP Beads by vortexing.
Transfer the DNA sample to a clean 1.5 ml Eppendorf DNA LoBind tube.
Add 60 µl of resuspended the AMPure XP Beads to the end-prep reaction and mix by flicking the tube.
Incubate on a Hula mixer (rotator mixer) for 5 minutes at room temperature.
Prepare 500 μl of fresh 80% ethanol in nuclease-free water.
Spin down the sample and pellet on a magnet until supernatant is clear and colourless. Keep the tube on the magnet, and pipette off the supernatant.
Keep the tube on the magnet and wash the beads with 200 µl of freshly prepared 80% ethanol without disturbing the pellet. Remove the ethanol using a pipette and discard.
Repeat the previous step.
Spin down and place the tube back on the magnet. Pipette off any residual ethanol. Allow to dry for ~30 seconds, but do not dry the pellet to the point of cracking.
Remove the tube from the magnetic rack and resuspend the pellet in 61 µl nuclease-free water. Incubate for 2 minutes at room temperature.
Pellet the beads on a magnet until the eluate is clear and colourless, for at least 1 minute.
Remove and retain 61 µl of eluate into a clean 1.5 ml Eppendorf DNA LoBind tube.
Quantify 1 µl of eluted sample using a Qubit fluorometer.
If a pause is required, the sample can be stored overnight at 4°C.
Although third-party ligase products may be supplied with their own buffer, the ligation efficiency of the Ligation Adapter (LA) is higher when using the Ligation Buffer (LNB) supplied in the Ligation Sequencing Kit.
Spin down the Ligation Adapter (LA) and Quick T4 Ligase, and place on ice.
Thaw Ligation Buffer (LNB) at room temperature, spin down and mix by pipetting. Due to viscosity, vortexing this buffer is ineffective. Place on ice immediately after thawing and mixing.
Thaw the Elution Buffer (EB) at room temperature and mix by vortexing. Then spin down and place on ice.
Thaw the Short Fragment Buffer (SFB) at room temperature and mix by vortexing. Then spin down and place on ice.
In a 1.5 ml Eppendorf DNA LoBind tube, mix in the following order:
Between each addition, pipette mix 10-20 times.
| Reagent | Volume |
|---|---|
| DNA sample from the previous step | 60 µl |
| Ligation Adapter (LA) | 5 µl |
| Ligation Buffer (LNB) | 25 µl |
| NEBNext Quick T4 DNA Ligase | 10 µl |
| Total | 100 µl |
Thoroughly mix the reaction by gently pipetting and briefly spinning down.
Incubate the reaction for 10 minutes at room temperature.
Resuspend the AMPure XP Beads by vortexing.
Add 40 µl of resuspended AMPure XP Beads to the reaction and mix by flicking the tube.
Incubate on a Hula mixer (rotator mixer) for 5 minutes at room temperature.
Spin down the sample and pellet on a magnet. Keep the tube on the magnet, and pipette off the supernatant when clear and colourless.
Wash the beads by adding 250 μl Short Fragment Buffer (SFB). Flick the beads to resuspend, spin down, then return the tube to the magnetic rack and allow the beads to pellet. Remove the supernatant using a pipette and discard.
Repeat the previous step.
Spin down and place the tube back on the magnet. Pipette off any residual supernatant. Allow to dry for ~30 seconds, but do not dry the pellet to the point of cracking.
Remove the tube from the magnetic rack and resuspend the pellet in 25 µl Elution Buffer (EB). Spin down and incubate for 10 minutes at room temperature. For high molecular weight DNA, incubating at 37°C can improve the recovery of long fragments.
Pellet the beads on a magnet until the eluate is clear and colourless, for at least 1 minute.
Remove and retain 25 µl of eluate containing the DNA library into a clean 1.5 ml Eppendorf DNA LoBind tube.
Dispose of the pelleted beads
Quantify 1 µl of eluted sample using a Qubit fluorometer.
Make up your library to 32 µl at 10-20 fmol, using Elution Buffer (EB).
We recommend loading 10-20 fmol of this final prepared library onto the R10.4.1 flow cell.
Loading more than 20 fmol of DNA can reduce the rate of duplex read capture. Dilute the library in Elution Buffer if required.
Take the 32 µl of the library forwards for loading onto the flow cell. Store on ice until ready to load.
10. Day 1: Priming and loading Duplex library on the PromethION Flow Cell
材料
- 测序缓冲液(SB)
- 文库颗粒(LIB)
- 文库溶液(LIS)
- 测序芯片系绳(FCT)
- 测序芯片冲洗液(FCF)
耗材
- PromethION 测序芯片(FLO-PRO114M)
- 1.5 ml Eppendorf DNA LoBind 离心管
仪器
- PromethION 测序设备
- PromethION 测序芯片遮光片
- P1000 移液枪和枪头
- P200 移液枪和枪头
- P20 移液枪和枪头
Day 1: Duplex experiment flow cell loading
Once the library has been prepared, the PromethION Flow Cell can be primed before the library is combined with the sequencing reagents and loaded into the flow cell.
We recommend monitoring your sequencing run and to reload your flow cell when recommended to in the "Washing and reloading Duplex library on PromethION Flow Cells". The Duplex experiment can be reloaded three times, across days 2, 3 and 4.
Using the Library Solution
For most sequencing experiments, use the Library Beads (LIB) for loading your library onto the flow cell. However, for viscous libraries it may be difficult to load with the beads and may be appropriate to load using the Library Solution (LIS).
Thaw the Sequencing Buffer (SB), Library Beads (LIB) or Library Solution (LIS, if using), Flow Cell Tether (FCT) and Flow Cell Flush (FCF) at room temperature before mixing by vortexing. Then spin down and store on ice.
Prepare the flow cell priming mix in a suitable tube for the number of flow cells to flush. Once combined, mix well by briefly vortexing.
| Reagents | Volume per flow cell |
|---|---|
| Flow Cell Flush (FCF) | 1,170 µl |
| Flow Cell Tether (FCT) | 30 µl |
| Total volume | 1,200 µl |
After taking flow cells out of the fridge, wait 20 minutes before inserting the flow cell into the PromethION for the flow cell to come to room temperature. Condensation can form on the flow cell in humid environments. Inspect the gold connector pins on the top and underside of the flow cell for condensation and wipe off with a lint-free wipe if any is observed. Ensure the heat pad (black pad) is present on the underside of the flow cell.
For the PromethION 24/48, load the flow cell(s) into the docking ports:
- Line up the flow cell with the connector horizontally and vertically before smoothly inserting into position.
- Press down firmly onto the flow cell and ensure the latch engages and clicks into place.


Insertion of the flow cells at the wrong angle can cause damage to the pins on the PromethION and affect your sequencing results. If you find the pins on a PromethION position are damaged, please contact support@nanoporetech.com for assistance.

Slide the inlet port cover clockwise to open.

Take care when drawing back buffer from the flow cell. Do not remove more than 20-30 µl, and make sure that the array of pores are covered by buffer at all times. Introducing air bubbles into the array can irreversibly damage pores.
After opening the inlet port, draw back a small volume to remove any air bubbles:
- Set a P1000 pipette tip to 200 µl.
- Insert the tip into the inlet port.
- Turn the wheel until the dial shows 220-230 µl, or until you see a small volume of buffer entering the pipette tip.

Load 500 µl of the priming mix into the flow cell via the inlet port, avoiding the introduction of air bubbles. Wait five minutes. During this time, prepare the library for loading using the next steps in the protocol.

Thoroughly mix the contents of the Library Beads (LIB) by pipetting.
The Library Beads (LIB) tube contains a suspension of beads. These beads settle very quickly. It is vital that they are mixed immediately before use.
We recommend using the Library Beads (LIB) for most sequencing experiments. However, the Library Solution (LIS) is available for more viscous libraries.
In a new 1.5 ml Eppendorf DNA LoBind tube, prepare the library for loading as follows:
| Reagent | Volume per flow cell |
|---|---|
| Sequencing Buffer (SB) | 100 µl |
| Library Beads (LIB) thoroughly mixed before use, or Library Solution (LIS) | 68 µl |
| DNA library | 32 µl |
| Total | 200 µl |
Note: Library loading volume has been increased to improve array coverage.
Complete the flow cell priming by slowly loading 500 µl of the priming mix into the inlet port.

Mix the prepared library gently by pipetting up and down just prior to loading.
Load 200 µl of library into the inlet port using a P1000 pipette.

Close the valve to seal the inlet port.
Install the light shield on your flow cell as soon as library has been loaded for optimal sequencing output.
We recommend leaving the light shield on the flow cell when library is loaded, including during any washing and reloading steps. The shield can be removed when the library has been removed from the flow cell.
If the light shield has been removed from the flow cell, install the light shield as follows:
- Align the inlet port cut out of the light shield with the inlet port cover on the flow cell. The leading edge of the light shield should sit above the flow cell ID.
- Firmly press the light shield around the inlet port cover. The inlet port clip will click into place underneath the inlet port cover.


Close the PromethION lid when ready to start a sequencing run on MinKNOW.
Wait a minimum of 10 minutes after loading the flow cells onto the PromethION before initiating any experiments. This will help to increase the sequencing output.
11. Day 2: Pore-C experiment
耗材
- T4 DNA Ligase 400,000 U/ml (NEB, M0202S/L)
- Recombinant Albumin at 20 μg/μl (NEB, B9200S)
- Tween-20 (Thermo Scientific, J20605.AP)
- Sodium dodecyl sulfate (SDS) at 10% v/v (Sigma, 71736)
- Proteinase K at 20 μg/μl (NEB, P8107S)
- 无核酸酶水(如ThermoFisher,AM9937)
- 1.5 ml Eppendorf DNA LoBind 离心管
- 2 ml Eppendorf DNA LoBind 离心管
仪器
- Thermomixer
- P1000 移液枪和枪头
- P200 移液枪和枪头
- P100 移液枪和枪头
- P20 移液枪和枪头
Day 2: Pore-C experiment overview
For Day 2, after the overnight incubation, the restriction enzymes are heat inactivated to prevent re-digesting ligated products. DNA ligase is added to the clusters of crosslinked DNA and passively diffuses through the crosslinked cytoskeleton cage to ligate the cohesive ends of proximal monomers into chimeric Pore-C polymers. After ligation, the ligated products can be released from the crosslinked cytoskeleton cages by an overnight proteinase K digestion. This releases the chimeric Pore-C polymers into solution as dsDNA.
Thaw the T4 DNA Ligase and T4 DNA Ligase Reaction Buffer in accordance with the manufacturer's instructions and place on ice.
- Thaw the reagents on ice.
- Flick and/or invert the reagent tube(s) to ensure they are well mixed.
Note: Do not vortex the T4 DNA Ligase enzyme. - Spin down tubes before opening for the first time each day.
Prepare 5 ml of 20% Tween-20 in nuclease free water as follows:
- Weigh out 1.095 g of Tween-20 and transfer to a fresh 5 ml centrifuge tube.
- Add 4 ml of nuclease-free water.
- Gently invert the tube until the solution is homogenous.
Set the thermomixer to 65°C.
Heat denature the restriction enzyme by incubating the sample suspension in the thermomixer at 65°C with 300 rpm rotation for 20 minutes. Allow the reaction to cool to room temperature.
Set the thermomixer to 16°C.
Set up the proximity ligation reaction according to the table below, adding reagents directly to the sample suspension in the following order. Mix gently by pipetting up and down, using a wide-bore pipette tip.
| Reagent | Final | Volume |
|---|---|---|
| Digestion reaction (from Day 1) | - | 450 µl |
| Nuclease-free water | - | 395 µl |
| T4 DNA Ligase Reaction Buffer, 10X | 1X | 100 µl |
| Recombinant albumin, 20 µg/µl | 0.1 µg/µl | 5 µl |
| T4 DNA Ligase, 400 U/µl | 20 U/µl | 50 µl |
| Total | - | 1000 µl |
Incubate the sample suspension in a thermomixer at 16°C for 6 hours, with periodic <1000 RPM rotation for <30 seconds every 15 minutes. This prevents condensation inside the lid.
Note: This incubation can be performed without mixing.
Do not extend incubation as prolonged ligation may increase trans-chromosomal contacts in the Pore-C data.
During this 6 hour incubation, the Ultra-long DNA experiment may be started.
Set the thermomixer to 56°C.
Add the reagents to the previous ligation reaction in the following order to make up the protein degradation reaction. Mix the sample gently by inverting the tube 3–4 times.
| Reagent | Final | Volume |
|---|---|---|
| Ligation reaction (from the Proximity Ligation) | - | 1000 μl |
| Nuclease-free water | - | 300 μl |
| Tween-20, 20% | 5% | 500 μl |
| SDS, 10% | 0.5% | 100 μl |
| Proteinase K, 20 μg/μl | 1 μg/μl | 100 μl |
| Total | - | 2000 μl |
Incubate the sample suspension in a thermomixer at 56°C for 18 hours with periodic <1000 rpm rotation for <30 seconds every 15 minutes to prevent condensation inside the lid.
Note: This incubation can be performed without mixing.
Incubation at 56°C compromises enzyme activity over a prolonged incubation. It is not advisable to incubate at higher temperatures as enzyme activity will reduce over time.
During the Pore-C incubation, start the Ultra-long DNA experiment.
12. Day 2: Ultra-long DNA experiment
材料
- 6 million PBMCs isolated from whole blood
- DNA 超长片段测序试剂盒(SQK-ULK114)
耗材
- Monarch® HMW DNA Extraction Kit for Tissue (NEB, T3060)
- Qubit dsDNA BR Assay(双链DNA宽范围检测)试剂盒(ThermoFisher ,Q32850)
- 磷酸盐缓冲盐溶液(PBS),pH 7.4(ThermoFisher,10010023)
- 异丙醇,100%(Fisher, 10723124)
- 乙醇,100%(例如 Fisher, 16606002)
- 5 ml Eppendorf DNA LoBind 离心管
- 2 ml Eppendorf DNA LoBind 离心管
- 1.5 ml Eppendorf DNA LoBind 离心管
仪器
- 热循环仪
- Temperature-controlled centrifuge
- 迷你离心机
- Hula混匀仪(低速旋转式混匀仪)
- 涡旋混匀仪
- Qubit荧光计 (或用于质控检测的等效仪器)
- P1000 移液枪和枪头
- P200 移液枪和枪头
- P20 移液枪和枪头
- 宽口移液枪头
- 盛有冰的冰桶
Day 2: Ultra-long DNA experiment overview
During the first day of the Ultra-long DNA experiment, the ultra-high molecular weight (uHMW) gDNA is extracted using the NEB Monarch HMW DNA Extraction Kit for Tissue. An optional uHMW gDNA quantification step has also been included. However, this step can be omitted and 750 µl of DNA in Extraction EB (EEB) can be taken straight into step 39 for tagmentation. During tagmentation, the DNA is cleaved and the transposase adapter simultaneously added to prepare the DNA ends for the attachment of the rapid adapter. The DNA is then cleaned using Precipitation Buffer (PTB) and incubated overnight.
Thaw the Extraction EB (EEB) at room temperature, mix by vortexing and place on ice.
Transfer 6 million cells resuspended in 40 µl PBS to a fresh 5 ml tube.
Thorough but gentle resuspension of cells is required to ensure efficient lysis and to prevent heterogeneity in the subsequent steps.
In a separate 2 ml Eppendorf DNA LoBind tube, combine the following reagents:
| Reagent | Volume |
|---|---|
| Monarch HMW gDNA Tissue Lysis Buffer | 1,800 µl |
| Proteinase K | 60 µl |
| Total | 1860 µl |
Add 1.8 ml of mixed Monarch HMW gDNA Tissue Lysis Buffer and Proteinase K to the resuspended cells.
Gently mix by slowly pipetting the reaction five times using a 1 ml wide-bore pipette tip.
Incubate the reaction at 56°C for 10 minutes.
Using a regular pipette tip, add 15 µl of Monarch RNase A.
Gently mix by slowly pipetting the reaction five times using a 1 ml wide-bore pipette tip.
Incubate the reaction at 56°C for 10 minutes on a thermomixer at 650 rpm.
Using a regular pipette tip, add 900 µl of the Monarch Protein Separation Solution to the reaction and mix using a Hula Mixer (rotator mixer) for 10 minutes, rotating at 3 rpm.
Centrifuge the reaction at 16,000 x g for 10 minutes at 4°C to separate the protein from the DNA.
DNA will be present in the upper phase, whereas protein and other contaminants will be in the lower phase.
Using a wide-bore pipette tip, carefully aspirate the upper phase containing the DNA and transfer to a fresh 5 ml tube without disturbing the phase below.
The DNA in the upper phase should be extremely viscous and should only be possible to aspirate using a wide-bore pipette tip.
If the protein phase is disturbed, the tube can be centrifuged again at 16,000 x g for 10 minutes at 4°C.
Add three Monarch DNA Capture Beads to the collected DNA phase.
Note: the first bead is sacrificial and will remain stuck at the bottom of the tube throughout the remainder of the process.
Add 2.5 ml isopropanol to the tube and mix using a Hula Mixer (rotator mixer) for 20 minutes rotating at 3 rpm. Ensure the DNA has fully precipitated around the glass beads.
Check the DNA is binding to the beads by looking for a viscous mass around the beads. The mixing step can be extended if the DNA is not obviously condensing around the beads.
Leave the tube to stand for 1 minute, without rotating, at room temperature.
Aspirate the supernatant from the tube, being careful not to aspirate the DNA that is bound to the beads. Check for and remove any supernatant remaining in the lid of the tube.
Note: if ~100 µl of supernatant is remaining in the tube, perfomance will not be affected.
Add 2 ml of Monarch gDNA Wash Buffer to the tube containing DNA bound to the beads and invert the tube to mix.
Ensure ethanol is added to the Monarch gDNA Wash Buffer as per kit guidance.
Aspirate the Wash Buffer, being careful not to aspirate the DNA that is bound to the beads. Check for and remove any Wash Buffer remaining in the lid of the tube.
Add 2 ml of Monarch gDNA Wash Buffer to the tube containing the DNA bound to the beads.
To a fresh 2 ml Eppendorf tube, add 560 µl of Extraction EB (EEB).
Aspirate the Wash Buffer, being careful not to aspirate the DNA that is bound to the beads. Check for and remove any Wash Buffer remaining in the lid of the tube.
Transfer the beads to a Monarch Bead Retainer inserted in a Monarch Collection Tube II.
Briefly spin the tube using a microfuge to remove any remaining Wash Buffer from the beads. Dispose of the collection tube containing residual wash buffer.
Do NOT use the Monarch Elution Buffer II in the Monarch® HMW DNA Extraction Kit for Tissue.
Immediately transfer the beads from the bead retainer into the 2 ml tube containing 560 µl of Extraction EB (EEB).
Beads should be transferred immediately to ensure that they do not over-dry, which could lead to increased solubilisation times.
Incubate the tube for 10 minutes at 56°C.
Pour the eluate and beads into a clean bead retainer inserted in a collection tube. Spin the tube at 1000 x g for 1 minute to separate eluate from the beads. Dispose of beads and bead retainer.
Add 200 µl of Extraction EB (EEB) to the collection tube to bring the total elution volume to 760 µl.
Transfer the eluate to a fresh 2 ml Eppendorf DNA LoBind tube.
Incubate the eluate for 10 minutes at 56°C.
Gently mix the eluate by slowly pipetting 10 times using a 1 ml wide-bore pipette tip.
Thorough but gentle resuspension of DNA is required to prevent heterogeneity in the sample.
At this point, the sample can be stored overnight at room temperature.
The next steps for DNA quantification are optional. Continue to the next stage of the protocol if quantification is to be omitted.
Use a regular P200 pipette tip to aspirate 10 µl of gDNA.
If the DNA is particularly viscous, the aspirated DNA can be separated from the sample by forcing the sample against the side of the tube to break the DNA off. It is critical that the DNA is completely homogenous, so that the 10 µl of sample that is removed is representative of the entire sample.
Dispense the aspirated gDNA into a fresh 2 ml Eppendorf DNA LoBind tube.
Add a Monarch DNA Capture Bead to the 10 µl of gDNA and vortex aggressively for 1 minute to shear the gDNA.
Transfer the gDNA and beads into a clean Monarch Bead Retainer inserted in a Monarch Collection Tube II. Spin the tube at 1000 x g for 1 minute to separate gDNA from the beads. Dispose of beads and bead retainer.
Transfer the gDNA into a clean 1.5 ml Eppendorf DNA LoBind tube.
Quantify the sample using a Qubit fluorometer. The expected yield is 30-40 µg of DNA.
Thaw the the kit components at room temperature, spin down briefly using a microfuge and mix by pipetting as indicated by the table below:
Once thawed, keep all the kit components on ice.
| Reagent | Thaw at room temperature | Briefly spin down | Mix well by pipetting |
|---|---|---|---|
| Fragmentation Mix (FRA) | Not frozen | ✓ | ✓ |
| FRA dilution buffer (FDB) | Not frozen | ✓ | ✓ |
| Rapid Adapter (RA) | Not frozen | ✓ | ✓ |
In a 1.5 ml Eppendorf DNA LoBind tube, dilute the Fragmentation Mix (FRA) with FRA Dilution Buffer (FDB) as follows:
| Reagent | Volume |
|---|---|
| Fragmentation Mix (FRA) | 6 µl |
| FRA dilution buffer (FDB) | 244 µl |
| Total | 250 µl |
Mix the diluted Fragmentation Mix (FRA) by pipetting.
Using a regular pipette tip, add 250 µl of diluted Fragmentation Mix (FRA) to the 750 µl of extracted DNA. Stir the reaction with the pipette tip whilst expelling the diluted Fragmentation Mix (FRA) to ensure an even distribution.
Immediately mix the reaction by slowly pipetting 10 times with a wide-bore pipette tip.
Visually check the reagents are thoroughly mixed. It is important to immediately mix the diluted Fragmentation Mix (FRA) with the DNA thoroughly.
Incubate the reaction as follows:
| Temperature | Time |
|---|---|
| Room temperature | 10 minutes |
| 75°C | 10 minutes |
| On ice | Cool on ice for a minimum of 10 minutes |
Note: the reaction must be cooled on ice before adding Rapid Adapter (RA) to prevent denaturing the enzyme.
Add 5 µl Rapid Adapter (RA) to the reaction using a regular pipette tip.
Gently mix the reaction by slowly pipetting five times using a 1 ml wide-bore pipette tip.
Note: visually check to ensure the reaction is thoroughly mixed.
Incubate the reaction for 30 minutes at room temperature.
Thaw the kit components at room temperature, spin down briefly using a microfuge and mix by vortexing as indicated by the table below:
| Reagent | Thaw at room temperature | Briefly spin down | Mix well by pipetting |
|---|---|---|---|
| Precipitation buffer (PTB) | ✓ | ✓ | ✓ |
| Elution Buffer (EB) | ✓ | ✓ | ✓ |
Once thawed, keep all the kit components on ice.
Using a regular pipette tip, add 500 µl of Precipitation Buffer (PTB) to the sample.
Mix the sample by rotating on a Hula Mixer (rotator mixer) for 20 minutes at 3 rpm.
Visually inspect to check the DNA has precipitated, forming a glassy white mass.
Centrifuge the sample at 1000 x g for 1 minute.
Using a regular pipette tip, carefully remove the supernatant from the tube, taking care not to aspirate the DNA pellet.
Centrifuge the sample at 1000 x g for 1 minute.
Using a regular pipette tip, carefully remove any residual supernatant from the tube, taking care not to aspirate the DNA pellet.
Using a regular pipette tip, add 300 µl of Elution Buffer (EB) to the tube containing the DNA. Incubate overnight at room temperature, for a minimum of 12 hours.
During the Ultra-long DNA experiment incubation, complete the first wash and reload of the Duplex experiment.
13. Day 2: Washing and reloading Duplex library on the PromethION Flow Cell
材料
- Flow Cell Wash Kit XL (EXP-WSH004-XL)
- 测序辅助扩展包 V14(EXP-AUX003)
- 测序缓冲液(SB)
- 文库颗粒(LIB)
- 文库溶液(LIS)
- 测序芯片系绳(FCT)
- 测序芯片冲洗液(FCF)
耗材
- 1.5 ml Eppendorf DNA LoBind 离心管
仪器
- P1000 移液枪和枪头
- P20 移液枪和枪头
- 盛有冰的冰桶
Day 2: Duplex experiment flow cell washing and reloading
We recommend washing and reloading your PromethION Flow Cell with a new library to maintain high data acquisition.
For the Duplex experiment, up to four libraries prepared using the Ligation Sequencing Kit XL V14 (SQK-LSK114-XL) can be loaded on the PromethION Flow Cell during a sequencing run. We recommend washing your flow cell when ~20-25% of active pores are remaining, which typically occurs after ~20-24 hours of sequencing. Washing removes most of the initial library as well as unblocking pores to prepare the flow cell for loading a new library for further sequencing.
Navigate to the Pore Activity or the Pore Scan Results plot to see pore availability. Below is an example of pore states observed on a flow cell before and after wash steps are performed. The red asterisks indicates the reloads. 
We recommend reloading your duplex experiment on days 2, 3 and 4 of the protocol.
Washing and reloading a PromethION Flow Cell video
This video will show you how to wash a flow cell after a sequencing run and how to load a new library.
We recommend keeping the light shield on the flow cell during washing if a second library will be loaded straight away.
If the flow cell is to be washed and stored, the light shield can be removed.
Place the tube of Wash Mix (WMX) on ice. Do not vortex the tube.
Thaw one tube of Wash Diluent (DIL) at room temperature.
Mix the contents of Wash Diluent (DIL) thoroughly by vortexing, then spin down briefly and place on ice.
In a fresh 1.5 ml Eppendorf DNA LoBind tube, prepare the following Flow Cell Wash Mix:
| Reagent | Volume per flow cell |
|---|---|
| Wash Mix (WMX) | 2 μl |
| Wash Diluent (DIL) | 398 μl |
| Total | 400 μl |
Mix well by pipetting, and place on ice. Do not vortex the tube.
Pause the sequencing experiment in MinKNOW, and leave the flow cell in the device.
It is vital that the inlet port is closed before removing waste to prevent air from being drawn across the sensor array area, which would lead to a significant loss of sequencing channels.
Remove waste buffer, as follows:
- Close the inlet port.
- Insert a P1000 pipette into a waste port and remove the waste buffer.
Note: As both the inlet port is closed, no fluid should leave the sensor array area.
Slide the inlet port cover clockwise to open the inlet port.

Take care when drawing back buffer from the flow cell. Do not remove more than 20-30 µl, and make sure that the array of pores are covered by buffer at all times. Introducing air bubbles into the array can irreversibly damage pores.
After opening the inlet port, check for a small air bubble under the cover. Draw back a small volume to remove any bubbles:
- Set a P1000 pipette to 200 µl
- Insert the tip into the inlet port
- Turn the wheel until the dial shows 220-230 µl, or until you can see a small volume of buffer entering the pipette tip.

Slowly load 200 µl of the prepared flow cell wash mix into the inlet port, as follows:
- Using a P1000 pipette, take 200 µl of the flow cell wash mix
- Insert the pipette tip into the inlet port, ensuring there are no bubbles in the tip
- Slowly twist the pipette wheel down to load the flow cell (if possible with your pipette) or push down the plunger very slowly, leaving a small volume of buffer in the pipette tip.
- Set a timer for a 5 minute incubation.
Close the inlet port and wait for 1 hour.
It is vital that the inlet port is closed before removing waste to prevent air from being drawn across the sensor array area, which would lead to a significant loss of sequencing channels.
Remove the waste buffer, as follows:
- Ensure the inlet port is closed.
- Insert a P1000 pipette into a waste port and remove the waste buffer
Note: As the inlet port is closed, no fluid should leave the sensor array area.
The buffers used in this process are incompatible with conducting a Flow Cell Check step prior to loading the subsequent library. However, number of available pores will be reported after the next pore scan.
Thaw the Sequencing Buffer (SB), Library Beads (LIB) or Library Solution (LIS, if using), Flow Cell Tether (FCT) and Flow Cell Flush (FCF) at room temperature before mixing by vortexing. Then spin down and store on ice.
Prepare the flow cell priming mix in a suitable tube for the number of flow cells to flush. Once combined, mix well by briefly vortexing.
| Reagents | Volume per flow cell |
|---|---|
| Flow Cell Flush (FCF) | 1,170 µl |
| Flow Cell Tether (FCT) | 30 µl |
| Total volume | 1,200 µl |
Slide the inlet port cover clockwise to open.

Take care when drawing back buffer from the flow cell. Do not remove more than 20-30 µl, and make sure that the array of pores are covered by buffer at all times. Introducing air bubbles into the array can irreversibly damage pores.
After opening the inlet port, draw back a small volume to remove any air bubbles:
- Set a P1000 pipette tip to 200 µl.
- Insert the tip into the inlet port.
- Turn the wheel until the dial shows 220-230 µl, or until you see a small volume of buffer entering the pipette tip.

Slowly load 500 µl of the priming mix into the inlet port, as follows:
- Using a P1000 pipette, take 500 µl of the priming mix
- Insert the pipette tip into the priming port, ensuring there are no bubbles in the tip
- Slowly twist the pipette wheel down to load the flow cell (if possible with your pipette) or push down the plunger very slowly, leaving a small volume of buffer in the pipette tip.

It is vital to wait five minutes between the priming mix flushes to ensure effective removal of the nuclease.
Close the inlet port and wait five minutes.
During this time, prepare the library for loading using the next steps in the protocol.
Thoroughly mix the contents of the Library Beads (LIB) by pipetting.
The Library Beads (LIB) tube contains a suspension of beads. These beads settle very quickly. It is vital that they are mixed immediately before use.
We recommend using the Library Beads (LIB) for most sequencing experiments. However, the Library Solution (LIS) is available for more viscous libraries.
In a new 1.5 ml Eppendorf DNA LoBind tube, prepare the library for loading as follows:
| Reagent | Volume per flow cell |
|---|---|
| Sequencing Buffer (SB) | 100 µl |
| Library Beads (LIB) thoroughly mixed before use, or Library Solution (LIS) | 68 µl |
| DNA library | 32 µl |
| Total | 200 µl |
Note: Library loading volume has been increased to improve array coverage.
It is vital that the inlet port is closed before removing waste to prevent air from being drawn across the sensor array area, which would lead to a significant loss of sequencing channels.
Remove the waste buffer, as follows:
- Ensure the inlet port is closed.
- Insert a P1000 pipette into a waste port and remove the waste buffer
Note: As the inlet port is closed, no fluid should leave the sensor array area.
Slide the inlet port cover clockwise to open.

Take care when drawing back buffer from the flow cell. Do not remove more than 20-30 µl, and make sure that the array of pores are covered by buffer at all times. Introducing air bubbles into the array can irreversibly damage pores.
After opening the inlet port, draw back a small volume to remove any air bubbles:
- Set a P1000 pipette tip to 200 µl.
- Insert the tip into the inlet port.
- Turn the wheel until the dial shows 220-230 µl, or until you see a small volume of buffer entering the pipette tip.

Slowly load 500 µl of the priming mix into the inlet port, as follows:
- Using a P1000 pipette, take 500 µl of the priming mix
- Insert the pipette tip into the priming port, ensuring there are no bubbles in the tip
- Slowly twist the pipette wheel down to load the flow cell (if possible with your pipette) or push down the plunger very slowly, leaving a small volume of buffer in the pipette tip.

It is vital that the inlet port is closed before removing waste to prevent air from being drawn across the sensor array area, which would lead to a significant loss of sequencing channels.
Remove waste buffer, as follows:
- Close the inlet port.
- Insert a P1000 pipette into a waste port and remove the waste buffer.
Note: As both the inlet port is closed, no fluid should leave the sensor array area.
Slide the inlet port cover clockwise to open.

Take care when drawing back buffer from the flow cell. Do not remove more than 20-30 µl, and make sure that the array of pores are covered by buffer at all times. Introducing air bubbles into the array can irreversibly damage pores.
After opening the inlet port, draw back a small volume to remove any air bubbles:
- Set a P1000 pipette tip to 200 µl.
- Insert the tip into the inlet port.
- Turn the wheel until the dial shows 220-230 µl, or until you see a small volume of buffer entering the pipette tip.

Mix the prepared library gently by pipetting up and down just prior to loading.
Load 200 µl of library into the inlet port using a P1000 pipette.

Close the valve to seal the inlet port.
Install the light shield on your flow cell as soon as library has been loaded for optimal sequencing output.
We recommend leaving the light shield on the flow cell when library is loaded, including during any washing and reloading steps. The shield can be removed when the library has been removed from the flow cell.
If the light shield has been removed from the flow cell, install the light shield as follows:
- Align the inlet port cut out of the light shield with the inlet port cover on the flow cell. The leading edge of the light shield should sit above the flow cell ID.
- Firmly press the light shield around the inlet port cover. The inlet port clip will click into place underneath the inlet port cover.


Close the PromethION lid when ready to start a sequencing run on MinKNOW.
Wait a minimum of 10 minutes after loading the flow cells onto the PromethION before initiating any experiments. This will help to increase the sequencing output.
Resume the sequencing run in MinKNOW to continue data acquisition.
14. Day 3: Pore-C experiment
材料
- Custom SPRI bead suspension
耗材
- Qubit dsDNA HS Assay(双链DNA高灵敏度检测)试剂盒(Invitrogen, Q32851)
- Chilled phenol:chloroform:isoamyl alcohol in a 25:24:1 ratio, saturated with 10 mM Tris.HCl pH 8.0, 1 mM EDTA (Sigma, P3803-400ML)
- 5 M NaCl (Sigma, 71386)
- 3 M sodium acetate, pH 5.5 (Invitrogen, AM9740)
- 0.5 M EDTA,pH 8(Thermo Scientific, R1021)
- 乙醇,100%(例如 Fisher, 16606002)
- TE 缓冲液(10 mM Tris-HCl、1 mM EDTA、pH 8.0)(Fisher scientific,10224683)
- Qubit™ 分析管(Invitrogen, Q32856)
- 5 ml centrifuge tubes
- 2 ml Eppendorf DNA LoBind 离心管
仪器
- Temperature-controlled centrifuge
- Hula混匀仪(低速旋转式混匀仪)
- 磁力架
- 热循环仪或恒温加热仪
- Qubit™ 荧光计(或用于质控检测的等效仪器)
- P1000 移液枪和枪头
- P200 移液枪和枪头
- P100 移液枪和枪头
- P20 移液枪和枪头
- -80°C freezer storage
Day 3: Pore-C experiment overview
On the third day of the Pore-C experiment, the chimeric Pore-C dsDNA polymers are purified from the solution of polypeptide fragments and residual reaction buffers. The peptides are removed by using a phenol:chloroform extraction, followed by an ethanol precipitation to purify the DNA from the residual reaction buffers and phenol. The final Pore-C DNA extract is a pool of chimeric dsDNA polymers made of multiple ligated monomers which are sequenced to determine DNA interactions, proximity in sequence space and the three-dimensional structures of chromatin within the nucleus.
Pre-cool the centrifuge to 15°C.
Place the sample on ice until cool, then transfer the entire volume to a 5 ml centrifuge tube.
Rinse the original tube with a further 200 μl of nuclease-free water and add this to the same 5 ml centrifuge tube for a total sample volume of ~2200 μl.
Add an equal volume of chilled phenol:chloroform:isoamyl alcohol 25:24:1 saturated with 10 mM Tris.HCl pH 8.0, 1 mM EDTA, adjusting this volume as needed to match that of the sample. Mix by gently inverting the tube for 5 minutes to achieve a homogeneous emulsion.
Centrifuge the aliquots at 16,000 x g at 15°C for 15 minutes.
Incubate the aliquots on ice for 2 minutes until the organic phase becomes cloudy; this will strengthen the integrity of the interphase layer.
If the protein degradation has been successful, the interphase layer will be very thin and clear.
Do not remove the interphase layer in the next step.
Transfer the aqueous phase into a fresh 5 ml centrifuge tube for each aliquot and make note of the recovered volume (expect ~2,000 μl).
Transfer half of the recovered aqueous phase to a second 5 ml centrifuge tube to create two equal aliquots.
For each aliquot, add 0.02X of 5 M NaCl (0.1 M final) and 0.1X of 3 M sodium acetate pH 5.5 (0.3 M final), relative to the volume of the recovered aqueous phase of the aliquot. Mix by gently inverting the tube.
The solution will likely turn cloudy and then become clear once again.
For example, for a recovered volume of 2,000 µl, add:
- 40 µl of 5 M NaCl
- 200 µl of 3 M sodium acetate
For each aliquot, add 3X of 100% ethanol relative to the volume of the recovered aqueous phase. Mix by gently inverting the tubes.
The solution will likely turn cloudy and then become clear once again.
Precipitate at –80°C for >1 hour.
Note: If a –80°C freezer is not available or a pause in the protocol is required, an overnight incubation at –20°C can be used instead.
Pre-cool a centrifuge to 4°C.
Centrifuge the sample at 16,000 x g at 4°C for 30 minutes.
Aspirate and discard the supernatant, then wash the pellets with 4 ml of 80% ethanol.
Centrifuge the sample at 16,000 x g at 4°C for 5 minutes.
Aspirate and discard the supernatant, then wash the pellets with 2 ml of 70% ethanol.
Centrifuge the sample at 16,000 x g at 4°C for 5 minutes.
Aspirate and discard the supernatant. Briefly spin down the tubes and aspirate any residual supernatant. Allow the pellets to dry for 5 minutes.
After the DNA pellets have dried, they may loosen from the tube.
Carefully resuspend each aliquot in 75 μl of TE buffer. Incubate for 5 minutes at room temperature, mixing by gently inverting the tube every few minutes.
Briefly spin down the tubes, then transfer and pool all aliquots together into a 1.5 ml Eppendorf DNA LoBind tube.
Quantify DNA concentration by using the Qubit dsDNA HS Assay Kit. Ensure a 1/10 dilution is used, as the Qubit reading will be affected by high salt concentration.
Note: The expected yield is ~7 μg per 10 million cells input for cell culture. Yields for other sample types may be reduced.
Dilute your sample to 60 ng/µl in a final volume of 50 µl of TE buffer at pH 8.
Add 42.5 µl (0.85X) of room temperature custom SPRI bead suspension and mix by flicking the tube.
Incubate on a Hula mixer (rotator mixer) for 10 minutes at room temperature.
Spin down briefly and pellet on a magnet until the supernatant is clear and colourless. Keep the tube on the magnet, and pipette off the supernatant.
Keep the tube on the magnet and wash the beads with 200 µl of freshly prepared 80% ethanol without disturbing the pellet. Remove the ethanol using a pipette and discard.
Repeat the previous step.
Spin down and place the tube back on the magnetic rack. Pipette off any residual ethanol. Allow the pellet to dry for ~30 seconds, but do not dry the pellet to the point of cracking.
Remove the tube from the magnetic rack and resuspend the pellet in 40 µl of TE buffer. Incubate for 1 minute at 50°C, and then for 5 minutes at room temperature.
Pellet the beads on a magnet until the eluate is clear and colourless, for at least 1 minute.
Remove and retain 40 µl of eluate into a clean 1.5 ml Eppendorf DNA LoBind tube.
Quantify 1 µl of eluted sample using a Qubit fluorometer.
You can expect a 50-55% loss of DNA depending on a fragment length distribution of input material: the greater the proportion of short fragments (<1.5-2 kb), the greater the sample loss.
Store the sample at 4°C and complete the Ultra-long DNA experiment.
The Pore-C sample will undergo library preparation during day 4.
15. Day 3: Ultra-long DNA experiment
耗材
- 1.5 ml Eppendorf DNA LoBind 离心管
仪器
- P1000 移液枪和枪头
- P100 移液枪和枪头
- P200 移液枪和枪头
- P20 移液枪和枪头
- 宽口移液枪头
Day 3: Ultra-long DNA experiment overview
The eluate from the overnight incubation is removed and stored on ice until it is ready for loading onto the PromethION Flow Cell.
Gently mix the DNA library by slowly pipetting 10 times with a wide-bore pipette tip.
Thorough but gentle resuspension of DNA is required to prevent heterogeneity in the sample.
Take the DNA library forwards for loading into the flow cell. Store the library on ice until ready to load.
16. Day 3: Priming and loading ultra-long DNA library on the PromethION Flow Cell
材料
- 测序芯片冲洗液(FCF)
- 冲洗系绳 UL(FTU)
- 上样溶液 UL(LSU)
- 测序缓冲液 UL(SBU)
耗材
- PromethION 测序芯片(FLO-PRO114M)
- 1.5 ml Eppendorf DNA LoBind 离心管
仪器
- PromethION 测序设备
- PromethION 测序芯片遮光片
- P1000 移液枪和枪头
- P200 移液枪和枪头
- P20 移液枪和枪头
- 宽口移液枪头
Day 3: Ultra-long DNA experiment flow cell loading
Once the ultra-long DNA library has been prepared, the PromethION Flow Cell can be primed and the library prepared with the final sequencing reagents before loading for sequencing to begin. Due to the viscosity of the library, the flow cell priming and loading steps have been modified.
We recommend monitoring your sequencing run and to reload your flow cell when recommended to in the "Washing and reloading the PromethION Flow Cell with ultra-long DNA library" section. The Ultra-long DNA experiment can be reloaded up to three times, across days 4 and 5.
Thaw the Sequencing Buffer UL (SBU), Loading Solution UL (LSU), Flush Tether UL (FTU) and one tube of Flow Cell Flush (FCF) at room temperature and mix by vortexing. Then spin down and place on ice.
In a fresh 1.5 ml Eppendorf DNA LoBind tube, prepare the DNA library for loading as follows using a wide-bore pipette tip for the addition of the DNA library:
| Reagent | Volume per flow cell |
|---|---|
| Sequencing Buffer UL (SBU) | 100 µl |
| Loading Solution UL (LSU) | 10 µl |
| DNA library | 90 µl |
| Total | 200 µl |
Note: ensure the Sequencing Buffer UL (SBU) and Loading Solution UL (LSU) are thoroughly mixed by pipetting before the addition of the DNA library.
Gently mix the prepared DNA library by slowly pipetting ten times using a wide-bore pipette tip.
Incubate at room temperature for 30 minutes then gently mix by slowly pipetting with a wide-bore tip. Visually inspect to ensure the sample is homogenous.
Prepare the flow cell priming mix in a 1.5 ml Eppendorf DNA LoBind tube and mix by vortexing at room temperature.
| Reagent | Volume |
|---|---|
| Flush Tether UL (FTU) | 30 µl |
| Flow Cell Flush (FCF) | 1170 µl |
| Total | 1200 µl |
After taking flow cells out of the fridge, wait 20 minutes before inserting the flow cell into the PromethION for the flow cell to come to room temperature. Condensation can form on the flow cell in humid environments. Inspect the gold connector pins on the top and underside of the flow cell for condensation and wipe off with a lint-free wipe if any is observed. Ensure the heat pad (black pad) is present on the underside of the flow cell.
For the PromethION 24/48, load the flow cell(s) into the docking ports:
- Line up the flow cell with the connector horizontally and vertically before smoothly inserting into position.
- Press down firmly onto the flow cell and ensure the latch engages and clicks into place.


Insertion of the flow cells at the wrong angle can cause damage to the pins on the PromethION and affect your sequencing results. If you find the pins on a PromethION position are damaged, please contact support@nanoporetech.com for assistance.

Slide the inlet port cover clockwise to open.

Take care when drawing back buffer from the flow cell. Do not remove more than 20-30 µl, and make sure that the array of pores are covered by buffer at all times. Introducing air bubbles into the array can irreversibly damage pores.
After opening the inlet port, draw back a small volume to remove any air bubbles:
- Set a P1000 pipette tip to 200 µl.
- Insert the tip into the inlet port.
- Turn the wheel until the dial shows 220-230 µl, or until you see a small volume of buffer entering the pipette tip.

Load 500 µl of the priming mix into the flow cell via the inlet port, avoiding the introduction of air bubbles. Wait five minutes.

Complete the flow cell priming by slowly loading 500 µl of the priming mix into the inlet port.

Ensure the inlet port cover of the flow cell is still open in preparation for loading.
Check that no air bubbles have been introduced to the inlet port during flow cell priming. If air is present, draw back a small volume to remove any air bubbles by using a P1000 pipette set to 200 µl and turning the pipette wheel (as per the instructions above).

Aspirate the DNA library with a wide-bore pipette tip. Ensure there are no air bubbles in the pipette tip. Place the wide-bore pipette tip directly on the inlet port. Slowly depress the pipette to dispense the library into the inlet port.
There can be a delay between depressing the pipette and the library dispensing from the pipette tip. Dispense the library slowly, allowing the library to dispense from the pipette tip before depressing the pipette further.
It is important to dispense the library slowly to prevent air being introduced onto the flow cell.
Note: The DNA library loaded in this step is viscous and may not readily flow through the inlet port into the flow cell. In this case, we recommend applying negative pressure in the flow cell as explained in the steps below.
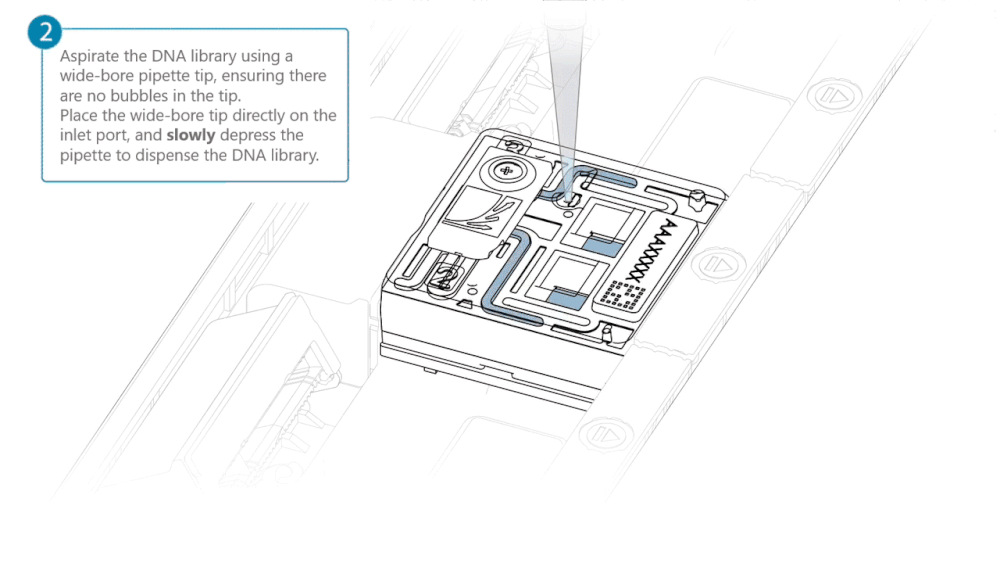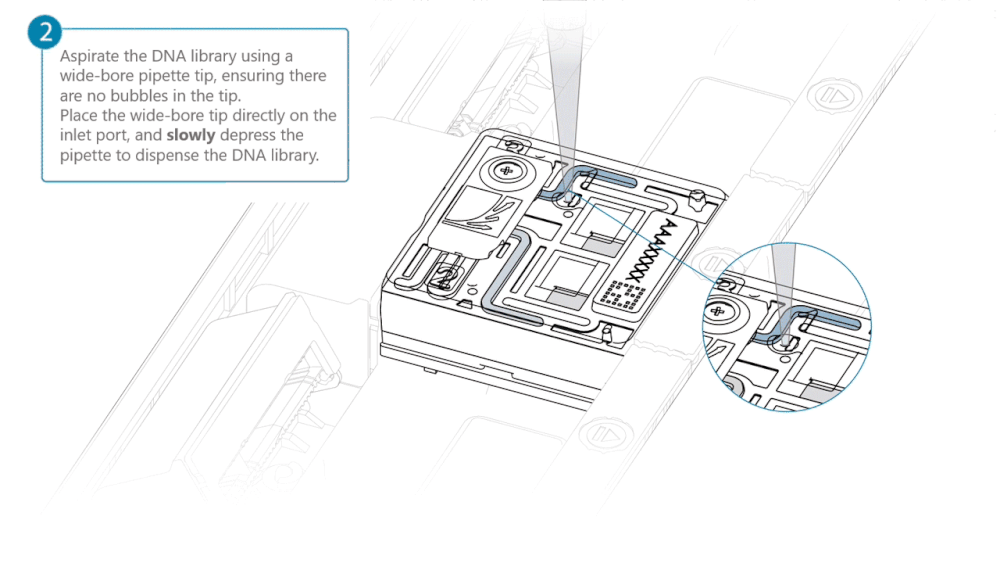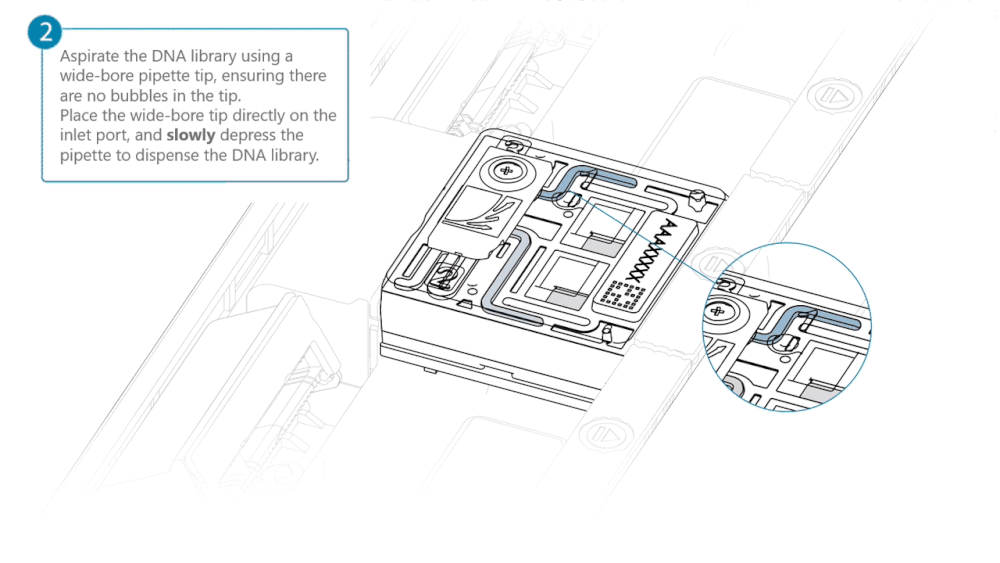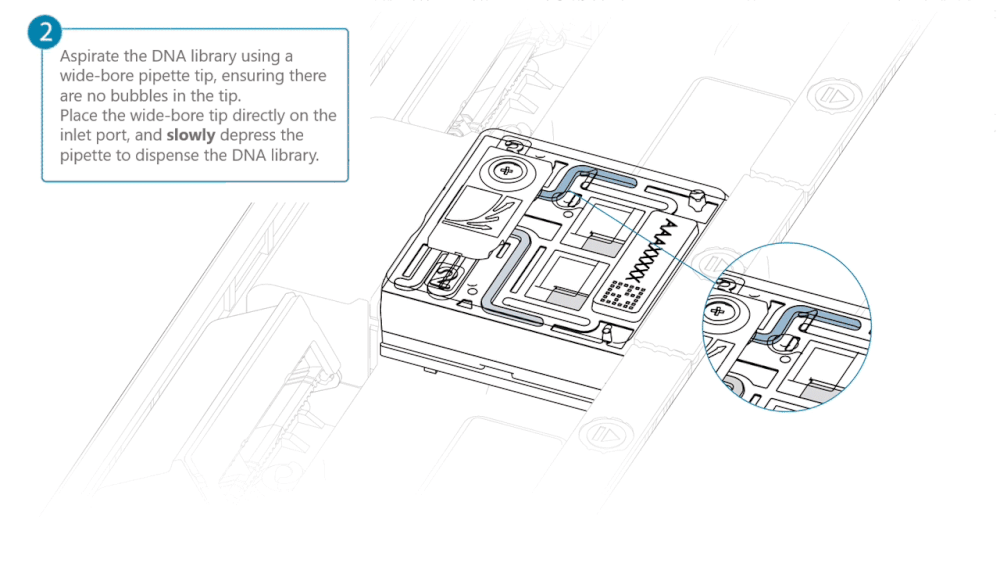
Using a P200 pipette, set the pipette to 50 µl and insert the tip into Port 2.

Very slowly turn the wheel of the pipette to pull the DNA library into the inlet port. Closely watch the DNA library on the inlet port and completely remove the pipette as soon as the library starts to be pulled into the port.
This step is required if the DNA library has not been fully absorbed into the inlet port.
Note: Take care to not apply too much negative pressure too quickly to avoid bringing air bubbles into the flow cell. Air bubbles will cause irreversible damage to the flow cell.

Close the valve to seal the inlet port.
Install the light shield on your flow cell as soon as library has been loaded for optimal sequencing output.
We recommend leaving the light shield on the flow cell when library is loaded, including during any washing and reloading steps. The shield can be removed when the library has been removed from the flow cell.
If the light shield has been removed from the flow cell, install the light shield as follows:
- Align the inlet port cut out of the light shield with the inlet port cover on the flow cell. The leading edge of the light shield should sit above the flow cell ID.
- Firmly press the light shield around the inlet port cover. The inlet port clip will click into place underneath the inlet port cover.


Close the PromethION lid when ready to start a sequencing run on MinKNOW.
Wait a minimum of 10 minutes after loading the flow cells onto the PromethION before initiating any experiments. This will help to increase the sequencing output.
17. Day 3: Washing and reloading Duplex library on the PromethION Flow Cell
材料
- Flow Cell Wash Kit XL (EXP-WSH004-XL)
- 测序辅助扩展包 V14(EXP-AUX003)
耗材
- 1.5 ml Eppendorf DNA LoBind 离心管
仪器
- P1000 移液枪和枪头
- P20 移液枪和枪头
- 盛有冰的冰桶
Day 3: Duplex experiment flow cell washing and reloading
We recommend washing and reloading your PromethION Flow Cell with a new library to maintain high data acquisition.
For the Duplex experiment, up to four libraries prepared using the Ligation Sequencing Kit XL V14 (SQK-LSK114-XL) can be loaded on the PromethION Flow Cell during a sequencing run. We recommend washing your flow cell when ~20-25% of active pores are remaining, which typically occurs after ~20-24 hours of sequencing. Washing removes most of the initial library as well as unblocking pores to prepare the flow cell for loading a new library for further sequencing.
Navigate to the Pore Activity or the Pore Scan Results plot to see pore availability. Below is an example of pore states observed on a flow cell before and after wash steps are performed. The red asterisks indicates the reloads. 
We recommend reloading your duplex experiment on days 2, 3 and 4 of the protocol.
Washing and reloading a PromethION Flow Cell video
This video will show you how to wash a flow cell after a sequencing run and how to load a new library.
We recommend keeping the light shield on the flow cell during washing if a second library will be loaded straight away.
If the flow cell is to be washed and stored, the light shield can be removed.
Place the tube of Wash Mix (WMX) on ice. Do not vortex the tube.
Thaw one tube of Wash Diluent (DIL) at room temperature.
Mix the contents of Wash Diluent (DIL) thoroughly by vortexing, then spin down briefly and place on ice.
In a fresh 1.5 ml Eppendorf DNA LoBind tube, prepare the following Flow Cell Wash Mix:
| Reagent | Volume per flow cell |
|---|---|
| Wash Mix (WMX) | 2 μl |
| Wash Diluent (DIL) | 398 μl |
| Total | 400 μl |
Mix well by pipetting, and place on ice. Do not vortex the tube.
Pause the sequencing experiment in MinKNOW, and leave the flow cell in the device.
It is vital that the inlet port is closed before removing waste to prevent air from being drawn across the sensor array area, which would lead to a significant loss of sequencing channels.
Remove waste buffer, as follows:
- Close the inlet port.
- Insert a P1000 pipette into a waste port and remove the waste buffer.
Note: As both the inlet port is closed, no fluid should leave the sensor array area.
Slide the inlet port cover clockwise to open the inlet port.

Take care when drawing back buffer from the flow cell. Do not remove more than 20-30 µl, and make sure that the array of pores are covered by buffer at all times. Introducing air bubbles into the array can irreversibly damage pores.
After opening the inlet port, draw back a small volume to remove any air bubbles:
- Set a P1000 pipette tip to 200 µl.
- Insert the tip into the inlet port.
- Turn the wheel until the dial shows 220-230 µl, or until you see a small volume of buffer entering the pipette tip.

Slowly load 200 µl of the prepared flow cell wash mix into the inlet port, as follows:
- Using a P1000 pipette, take 200 µl of the flow cell wash mix
- Insert the pipette tip into the inlet port, ensuring there are no bubbles in the tip
- Slowly twist the pipette wheel down to load the flow cell (if possible with your pipette) or push down the plunger very slowly, leaving a small volume of buffer in the pipette tip.
- Set a timer for a 5 minute incubation.
Close the inlet port and wait for 1 hour.
It is vital that the inlet port is closed before removing waste to prevent air from being drawn across the sensor array area, which would lead to a significant loss of sequencing channels.
Remove the waste buffer, as follows:
- Ensure the inlet port is closed.
- Insert a P1000 pipette into a waste port and remove the waste buffer
Note: As the inlet port is closed, no fluid should leave the sensor array area.
The buffers used in this process are incompatible with conducting a Flow Cell Check step prior to loading the subsequent library. However, number of available pores will be reported after the next pore scan.
Thaw the Sequencing Buffer (SB), Library Beads (LIB) or Library Solution (LIS, if using), Flow Cell Tether (FCT) and Flow Cell Flush (FCF) at room temperature before mixing by vortexing. Then spin down and store on ice.
Prepare the flow cell priming mix in a suitable tube for the number of flow cells to flush. Once combined, mix well by briefly vortexing.
| Reagents | Volume per flow cell |
|---|---|
| Flow Cell Flush (FCF) | 1,170 µl |
| Flow Cell Tether (FCT) | 30 µl |
| Total volume | 1,200 µl |
Slide the inlet port cover clockwise to open the inlet port.

Take care when drawing back buffer from the flow cell. Do not remove more than 20-30 µl, and make sure that the array of pores are covered by buffer at all times. Introducing air bubbles into the array can irreversibly damage pores.
After opening the inlet port, draw back a small volume to remove any air bubbles:
- Set a P1000 pipette tip to 200 µl.
- Insert the tip into the inlet port.
- Turn the wheel until the dial shows 220-230 µl, or until you see a small volume of buffer entering the pipette tip.

Slowly load 500 µl of the priming mix into the inlet port, as follows:
- Using a P1000 pipette, take 500 µl of the priming mix
- Insert the pipette tip into the priming port, ensuring there are no bubbles in the tip
- Slowly twist the pipette wheel down to load the flow cell (if possible with your pipette) or push down the plunger very slowly, leaving a small volume of buffer in the pipette tip.

It is vital to wait five minutes between the priming mix flushes to ensure effective removal of the nuclease.
Close the inlet port and wait five minutes.
During this time, prepare the library for loading using the next steps in the protocol.
Thoroughly mix the contents of the Library Beads (LIB) by pipetting.
The Library Beads (LIB) tube contains a suspension of beads. These beads settle very quickly. It is vital that they are mixed immediately before use.
We recommend using the Library Beads (LIB) for most sequencing experiments. However, the Library Solution (LIS) is available for more viscous libraries.
In a new 1.5 ml Eppendorf DNA LoBind tube, prepare the library for loading as follows:
| Reagent | Volume per flow cell |
|---|---|
| Sequencing Buffer (SB) | 100 µl |
| Library Beads (LIB) thoroughly mixed before use, or Library Solution (LIS) | 68 µl |
| DNA library | 32 µl |
| Total | 200 µl |
Note: Library loading volume has been increased to improve array coverage.
It is vital that the inlet port is closed before removing waste to prevent air from being drawn across the sensor array area, which would lead to a significant loss of sequencing channels.
Remove the waste buffer, as follows:
- Ensure the inlet port is closed.
- Insert a P1000 pipette into a waste port and remove the waste buffer
Note: As the inlet port is closed, no fluid should leave the sensor array area.
Slide the inlet port cover clockwise to open.

Take care when drawing back buffer from the flow cell. Do not remove more than 20-30 µl, and make sure that the array of pores are covered by buffer at all times. Introducing air bubbles into the array can irreversibly damage pores.
After opening the inlet port, draw back a small volume to remove any air bubbles:
- Set a P1000 pipette tip to 200 µl.
- Insert the tip into the inlet port.
- Turn the wheel until the dial shows 220-230 µl, or until you see a small volume of buffer entering the pipette tip.

Slowly load 500 µl of the priming mix into the inlet port, as follows:
- Using a P1000 pipette, take 500 µl of the priming mix
- Insert the pipette tip into the priming port, ensuring there are no bubbles in the tip
- Slowly twist the pipette wheel down to load the flow cell (if possible with your pipette) or push down the plunger very slowly, leaving a small volume of buffer in the pipette tip.

It is vital that the inlet port is closed before removing waste to prevent air from being drawn across the sensor array area, which would lead to a significant loss of sequencing channels.
Remove waste buffer, as follows:
- Close the inlet port.
- Insert a P1000 pipette into a waste port and remove the waste buffer.
Note: As both the inlet port is closed, no fluid should leave the sensor array area.
Slide the inlet port cover clockwise to open the inlet port.

Take care when drawing back buffer from the flow cell. Do not remove more than 20-30 µl, and make sure that the array of pores are covered by buffer at all times. Introducing air bubbles into the array can irreversibly damage pores.
After opening the inlet port, draw back a small volume to remove any air bubbles:
- Set a P1000 pipette tip to 200 µl.
- Insert the tip into the inlet port.
- Turn the wheel until the dial shows 220-230 µl, or until you see a small volume of buffer entering the pipette tip.

Mix the prepared library gently by pipetting up and down just prior to loading.
Load 200 µl of library into the inlet port using a P1000 pipette.

Close the valve to seal the inlet port.
Install the light shield on your flow cell as soon as library has been loaded for optimal sequencing output.
We recommend leaving the light shield on the flow cell when library is loaded, including during any washing and reloading steps. The shield can be removed when the library has been removed from the flow cell.
If the light shield has been removed from the flow cell, install the light shield as follows:
- Align the inlet port cut out of the light shield with the inlet port cover on the flow cell. The leading edge of the light shield should sit above the flow cell ID.
- Firmly press the light shield around the inlet port cover. The inlet port clip will click into place underneath the inlet port cover.


Close the PromethION lid when ready to start a sequencing run on MinKNOW.
Wait a minimum of 10 minutes after loading the flow cells onto the PromethION before initiating any experiments. This will help to increase the sequencing output.
Resume the sequencing run in MinKNOW to continue data acquisition.
18. Day 4: Pore-C experiment
材料
- 100-200 fmol Pore-C DNA extract
- Ligation Sequencing Kit XL V14 (SQK-LSK114-XL)
耗材
- NEBNext FFPE DNA 修复混合液(NEB,M6630)
- NEBNext® Ultra II 末端修复/ dA尾添加模块(NEB,E7546)
- NEBNext 快速连接模块(NEB,E6056)
- Qubit dsDNA HS Assay(双链DNA高灵敏度检测)试剂盒(Invitrogen, Q32851)
- Agencourt AMPure XP 磁珠(Beckman Coulter™, A63881)
- 无核酸酶水(如ThermoFisher,AM9937)
- 新制备的 80% 乙醇(用无核酸酶水配制)
- Qubit™ 分析管(Invitrogen, Q32856)
- 1.5 ml Eppendorf DNA LoBind 离心管
- 0.2 ml 薄壁PCR管
仪器
- P1000 移液枪和枪头
- P100 移液枪和枪头
- P10 移液枪和枪头
- 热循环仪
- 迷你离心机
- Hula混匀仪(低速旋转式混匀仪)
- 磁力架
- 涡旋混匀仪
- 盛有冰的冰桶
- Qubit荧光计 (或用于质控检测的等效仪器)
Day 4: Pore-C experiment overview
The Pore-C DNA extracts are prepared for sequencing using the Ligation Sequencing Kit XL V14 (SQK-LSK114-XL), as follows: the extracted DNA is repaired and the ends prepared for adapter attachment using the NEBNext FFPE DNA Repair Mix and NEBNext Ultra II End Repair/dA-tailing Module reagents. The sequencing adapters are attached to the DNA fragment ends before a clean-up step in preparation for sequencing on the PromethION Flow Cell.
Thaw DNA Control Sample (DCS) at room temperature, spin down, mix by pipetting, and place on ice.
We recommend using the DNA Control Sample (DCS) in your library prep for troubleshooting purposes. However, you can omit this step and make up the extra 1 µl with your sample DNA.
Prepare the NEBNext FFPE DNA Repair Mix and NEBNext Ultra II End Repair / dA-tailing Module reagents in accordance with manufacturer’s instructions, and place on ice.
For optimal performance, NEB recommend the following:
Thaw all reagents on ice.
Flick and/or invert the reagent tubes to ensure they are well mixed.
Note: Do not vortex the FFPE DNA Repair Mix or Ultra II End Prep Enzyme Mix.Always spin down tubes before opening for the first time each day.
The Ultra II End Prep Buffer and FFPE DNA Repair Buffer may have a little precipitate. Allow the mixture to come to room temperature and pipette the buffer up and down several times to break up the precipitate, followed by vortexing the tube for 30 seconds to solubilise any precipitate.
Note: It is important the buffers are mixed well by vortexing.The FFPE DNA Repair Buffer may have a yellow tinge and is fine to use if yellow.
Prepare the DNA in nuclease-free water:
- Transfer 100-200 fmol input DNA into a 1.5 ml Eppendorf DNA LoBind tube.
- Adjust the volume to 47 μl with nuclease-free water.
- Mix thoroughly by pipetting up and down, or by flicking the tube.
- Spin down briefly in a microfuge
In a 0.2 ml thin-walled PCR tube, mix the following:
Between each addition, pipette mix 10-20 times.
| Reagent | Volume |
|---|---|
| DNA from the previous step | 47 µl |
| DNA CS (optional) | 1 µl |
| NEBNext FFPE DNA Repair Buffer | 3.5 µl |
| NEBNext FFPE DNA Repair Mix | 2 µl |
| Ultra II End-prep Reaction Buffer | 3.5 µl |
| Ultra II End-prep Enzyme Mix | 3 µl |
| Total | 60 µl |
Thoroughly mix the reaction by gently pipetting and briefly spinning down.
Using a thermal cycler, incubate at 20°C for 15 minutes and 65°C for 5 minutes.
Resuspend the AMPure XP Beads by vortexing.
Transfer the DNA sample to a clean 1.5 ml Eppendorf DNA LoBind tube.
Add 60 µl of resuspended the AMPure XP Beads to the end-prep reaction and mix by flicking the tube.
Incubate on a Hula mixer (rotator mixer) for 5 minutes at room temperature.
Prepare 500 μl of fresh 80% ethanol in nuclease-free water.
Spin down the sample and pellet on a magnet until supernatant is clear and colourless. Keep the tube on the magnet, and pipette off the supernatant.
Keep the tube on the magnet and wash the beads with 200 µl of freshly prepared 80% ethanol without disturbing the pellet. Remove the ethanol using a pipette and discard.
Repeat the previous step.
Spin down and place the tube back on the magnet. Pipette off any residual ethanol. Allow to dry for ~30 seconds, but do not dry the pellet to the point of cracking.
Remove the tube from the magnetic rack and resuspend the pellet in 61 µl nuclease-free water. Incubate for 2 minutes at room temperature.
Pellet the beads on a magnet until the eluate is clear and colourless, for at least 1 minute.
Remove and retain 61 µl of eluate into a clean 1.5 ml Eppendorf DNA LoBind tube.
Quantify 1 µl of eluted sample using a Qubit fluorometer.
If a pause is required, the sample can be stored overnight at 4°C.
Although third-party ligase products may be supplied with their own buffer, the ligation efficiency of the Ligation Adapter (LA) is higher when using the Ligation Buffer (LNB) supplied in the Ligation Sequencing Kit.
Spin down the Ligation Adapter (LA) and Quick T4 Ligase, and place on ice.
Thaw Ligation Buffer (LNB) at room temperature, spin down and mix by pipetting. Due to viscosity, vortexing this buffer is ineffective. Place on ice immediately after thawing and mixing.
Thaw the Elution Buffer (EB) at room temperature and mix by vortexing. Then spin down and place on ice.
Thaw the Short Fragment Buffer (SFB) at room temperature and mix by vortexing. Then spin down and place on ice.
In a 1.5 ml Eppendorf DNA LoBind tube, mix in the following order:
Between each addition, pipette mix 10-20 times.
| Reagent | Volume |
|---|---|
| DNA sample from the previous step | 60 µl |
| Ligation Adapter (LA) | 5 µl |
| Ligation Buffer (LNB) | 25 µl |
| NEBNext Quick T4 DNA Ligase | 10 µl |
| Total | 100 µl |
Thoroughly mix the reaction by gently pipetting and briefly spinning down.
Incubate the reaction for 10 minutes at room temperature.
Resuspend the AMPure XP Beads by vortexing.
Add 40 µl of resuspended AMPure XP Beads to the reaction and mix by flicking the tube.
Incubate on a Hula mixer (rotator mixer) for 5 minutes at room temperature.
Spin down the sample and pellet on a magnet. Keep the tube on the magnet, and pipette off the supernatant when clear and colourless.
Wash the beads by adding 250 μl Short Fragment Buffer (SFB). Flick the beads to resuspend, spin down, then return the tube to the magnetic rack and allow the beads to pellet. Remove the supernatant using a pipette and discard.
Repeat the previous step.
Spin down and place the tube back on the magnet. Pipette off any residual supernatant. Allow to dry for ~30 seconds, but do not dry the pellet to the point of cracking.
Remove the tube from the magnetic rack and resuspend the pellet in 25 µl Elution Buffer (EB). Spin down and incubate for 10 minutes at room temperature. For high molecular weight DNA, incubating at 37°C can improve the recovery of long fragments.
Pellet the beads on a magnet until the eluate is clear and colourless, for at least 1 minute.
Remove and retain 25 µl of eluate containing the DNA library into a clean 1.5 ml Eppendorf DNA LoBind tube.
Dispose of the pelleted beads
Quantify 1 µl of eluted sample using a Qubit fluorometer.
Make up your library to 32 µl at 10-20 fmol, using Elution Buffer (EB).
We recommend loading 10-20 fmol of this final prepared library onto the R10.4.1 flow cell.
Loading more than 20 fmol of DNA can reduce the rate of duplex read capture. Dilute the library in Elution Buffer if required.
The prepared library is used for loading onto the flow cell. Store the library on ice until ready to load.
19. Day 4: Priming and loading Pore-C library on the PromethION Flow Cell
材料
- 测序缓冲液(SB)
- 文库颗粒(LIB)
- 文库溶液(LIS)
- 测序芯片系绳(FCT)
- 测序芯片冲洗液(FCF)
耗材
- PromethION 测序芯片(FLO-PRO114M)
- 1.5 ml Eppendorf DNA LoBind 离心管
仪器
- PromethION 测序设备
- PromethION 测序芯片遮光片
- P1000 移液枪和枪头
- P200 移液枪和枪头
- P20 移液枪和枪头
Day 4: Pore-C experiment flow cell loading
Once the Pore-C DNA extracts have been prepared using the Ligation Sequencing Kit XL V14 (SQK-LSK114-XL), the PromethION Flow Cell can be primed and the library prepared with the final sequencing reagents for the first library load to be sequenced.
We recommend monitoring your sequencing run and to reload your flow cell when recommended to in the "Washing and reloading Pore-C library on the PromethION Flow Cell" section. Pore-C DNA extracts are more prone to cause flow cell blocking and may require washing and reloading earlier during a sequencing run compared to the duplex condition. The Pore-C condition can be reloaded three times, across days 5, 6 and 7.
Using the Library Solution
For most sequencing experiments, use the Library Beads (LIB) for loading your library onto the flow cell. However, for viscous libraries it may be difficult to load with the beads and may be appropriate to load using the Library Solution (LIS).
Thaw the Sequencing Buffer (SB), Library Beads (LIB) or Library Solution (LIS, if using), Flow Cell Tether (FCT) and Flow Cell Flush (FCF) at room temperature before mixing by vortexing. Then spin down and store on ice.
Prepare the flow cell priming mix in a suitable tube for the number of flow cells to flush. Once combined, mix well by briefly vortexing.
| Reagents | Volume per flow cell |
|---|---|
| Flow Cell Flush (FCF) | 1,170 µl |
| Flow Cell Tether (FCT) | 30 µl |
| Total volume | 1,200 µl |
After taking flow cells out of the fridge, wait 20 minutes before inserting the flow cell into the PromethION for the flow cell to come to room temperature. Condensation can form on the flow cell in humid environments. Inspect the gold connector pins on the top and underside of the flow cell for condensation and wipe off with a lint-free wipe if any is observed. Ensure the heat pad (black pad) is present on the underside of the flow cell.
For the PromethION 24/48, load the flow cell(s) into the docking ports:
- Line up the flow cell with the connector horizontally and vertically before smoothly inserting into position.
- Press down firmly onto the flow cell and ensure the latch engages and clicks into place.


Insertion of the flow cells at the wrong angle can cause damage to the pins on the PromethION and affect your sequencing results. If you find the pins on a PromethION position are damaged, please contact support@nanoporetech.com for assistance.

Slide the inlet port cover clockwise to open.

Take care when drawing back buffer from the flow cell. Do not remove more than 20-30 µl, and make sure that the array of pores are covered by buffer at all times. Introducing air bubbles into the array can irreversibly damage pores.
After opening the inlet port, draw back a small volume to remove any air bubbles:
- Set a P1000 pipette tip to 200 µl.
- Insert the tip into the inlet port.
- Turn the wheel until the dial shows 220-230 µl, or until you see a small volume of buffer entering the pipette tip.

Load 500 µl of the priming mix into the flow cell via the inlet port, avoiding the introduction of air bubbles. Wait five minutes. During this time, prepare the library for loading using the next steps in the protocol.

Thoroughly mix the contents of the Library Beads (LIB) by pipetting.
The Library Beads (LIB) tube contains a suspension of beads. These beads settle very quickly. It is vital that they are mixed immediately before use.
We recommend using the Library Beads (LIB) for most sequencing experiments. However, the Library Solution (LIS) is available for more viscous libraries.
In a new 1.5 ml Eppendorf DNA LoBind tube, prepare the library for loading as follows:
| Reagent | Volume per flow cell |
|---|---|
| Sequencing Buffer (SB) | 100 µl |
| Library Beads (LIB) thoroughly mixed before use, or Library Solution (LIS) | 68 µl |
| DNA library | 32 µl |
| Total | 200 µl |
Note: Library loading volume has been increased to improve array coverage.
Complete the flow cell priming by slowly loading 500 µl of the priming mix into the inlet port.

Mix the prepared library gently by pipetting up and down just prior to loading.
Load 200 µl of library into the inlet port using a P1000 pipette.

Close the valve to seal the inlet port.
Install the light shield on your flow cell as soon as library has been loaded for optimal sequencing output.
We recommend leaving the light shield on the flow cell when library is loaded, including during any washing and reloading steps. The shield can be removed when the library has been removed from the flow cell.
If the light shield has been removed from the flow cell, install the light shield as follows:
- Align the inlet port cut out of the light shield with the inlet port cover on the flow cell. The leading edge of the light shield should sit above the flow cell ID.
- Firmly press the light shield around the inlet port cover. The inlet port clip will click into place underneath the inlet port cover.


Close the PromethION lid when ready to start a sequencing run on MinKNOW.
Wait a minimum of 10 minutes after loading the flow cells onto the PromethION before initiating any experiments. This will help to increase the sequencing output.
20. Day 4: Washing and reloading Duplex library on the PromethION Flow Cell
材料
- Flow Cell Wash Kit XL (EXP-WSH004-XL)
- 测序辅助扩展包 V14(EXP-AUX003)
- 测序缓冲液(SB)
- 文库颗粒(LIB)
- 文库溶液(LIS)
- 测序芯片系绳(FCT)
- 测序芯片冲洗液(FCF)
耗材
- 1.5 ml Eppendorf DNA LoBind 离心管
仪器
- P1000 移液枪和枪头
- P20 移液枪和枪头
- 盛有冰的冰桶
Day 4: Duplex experiment flow cell washing and reloading
We recommend washing and reloading your PromethION Flow Cell with a new library to maintain high data acquisition.
For the Duplex experiment, up to four libraries prepared using the Ligation Sequencing Kit XL V14 (SQK-LSK114-XL) can be loaded on the PromethION Flow Cell during a sequencing run. We recommend washing your flow cell when ~20-25% of active pores are remaining, which typically occurs after ~20-24 hours of sequencing. Washing removes most of the initial library as well as unblocking pores to prepare the flow cell for loading a new library for further sequencing.
Navigate to the Pore Activity or the Pore Scan Results plot to see pore availability. Below is an example of pore states observed on a flow cell before and after wash steps are performed. The red asterisks indicates the reloads. 
We recommend reloading your Duplex experiment on days 2, 3 and 4 of the protocol.
Washing and reloading a PromethION Flow Cell video
This video will show you how to wash a flow cell after a sequencing run and how to load a new library.
We recommend keeping the light shield on the flow cell during washing if a second library will be loaded straight away.
If the flow cell is to be washed and stored, the light shield can be removed.
Place the tube of Wash Mix (WMX) on ice. Do not vortex the tube.
Thaw one tube of Wash Diluent (DIL) at room temperature.
Mix the contents of Wash Diluent (DIL) thoroughly by vortexing, then spin down briefly and place on ice.
In a fresh 1.5 ml Eppendorf DNA LoBind tube, prepare the following Flow Cell Wash Mix:
| Reagent | Volume per flow cell |
|---|---|
| Wash Mix (WMX) | 2 μl |
| Wash Diluent (DIL) | 398 μl |
| Total | 400 μl |
Mix well by pipetting, and place on ice. Do not vortex the tube.
Pause the sequencing experiment in MinKNOW, and leave the flow cell in the device.
It is vital that the inlet port is closed before removing waste to prevent air from being drawn across the sensor array area, which would lead to a significant loss of sequencing channels.
Remove waste buffer, as follows:
- Close the inlet port.
- Insert a P1000 pipette into a waste port and remove the waste buffer.
Note: As both the inlet port is closed, no fluid should leave the sensor array area.
Slide the inlet port cover clockwise to open the inlet port.

Take care when drawing back buffer from the flow cell. Do not remove more than 20-30 µl, and make sure that the array of pores are covered by buffer at all times. Introducing air bubbles into the array can irreversibly damage pores.
After opening the inlet port, check for a small air bubble under the cover. Draw back a small volume to remove any bubbles:
- Set a P1000 pipette to 200 µl
- Insert the tip into the inlet port
- Turn the wheel until the dial shows 220-230 µl, or until you can see a small volume of buffer entering the pipette tip.

Slowly load 200 µl of the prepared flow cell wash mix into the inlet port, as follows:
- Using a P1000 pipette, take 200 µl of the flow cell wash mix
- Insert the pipette tip into the inlet port, ensuring there are no bubbles in the tip
- Slowly twist the pipette wheel down to load the flow cell (if possible with your pipette) or push down the plunger very slowly, leaving a small volume of buffer in the pipette tip.
- Set a timer for a 5 minute incubation.
Close the inlet port and wait for 1 hour.
It is vital that the inlet port is closed before removing waste to prevent air from being drawn across the sensor array area, which would lead to a significant loss of sequencing channels.
Remove the waste buffer, as follows:
- Ensure the inlet port is closed.
- Insert a P1000 pipette into a waste port and remove the waste buffer
Note: As the inlet port is closed, no fluid should leave the sensor array area.
The buffers used in this process are incompatible with conducting a Flow Cell Check step prior to loading the subsequent library. However, number of available pores will be reported after the next pore scan.
Thaw the Sequencing Buffer (SB), Library Beads (LIB) or Library Solution (LIS, if using), Flow Cell Tether (FCT) and Flow Cell Flush (FCF) at room temperature before mixing by vortexing. Then spin down and store on ice.
Prepare the flow cell priming mix in a suitable tube for the number of flow cells to flush. Once combined, mix well by briefly vortexing.
| Reagents | Volume per flow cell |
|---|---|
| Flow Cell Flush (FCF) | 1,170 µl |
| Flow Cell Tether (FCT) | 30 µl |
| Total volume | 1,200 µl |
Slide the inlet port cover clockwise to open.

Take care when drawing back buffer from the flow cell. Do not remove more than 20-30 µl, and make sure that the array of pores are covered by buffer at all times. Introducing air bubbles into the array can irreversibly damage pores.
After opening the inlet port, draw back a small volume to remove any air bubbles:
- Set a P1000 pipette tip to 200 µl.
- Insert the tip into the inlet port.
- Turn the wheel until the dial shows 220-230 µl, or until you see a small volume of buffer entering the pipette tip.

Slowly load 500 µl of the priming mix into the inlet port, as follows:
- Using a P1000 pipette, take 500 µl of the priming mix
- Insert the pipette tip into the priming port, ensuring there are no bubbles in the tip
- Slowly twist the pipette wheel down to load the flow cell (if possible with your pipette) or push down the plunger very slowly, leaving a small volume of buffer in the pipette tip.

It is vital to wait five minutes between the priming mix flushes to ensure effective removal of the nuclease.
Close the inlet port and wait five minutes.
During this time, prepare the library for loading using the next steps in the protocol.
Thoroughly mix the contents of the Library Beads (LIB) by pipetting.
The Library Beads (LIB) tube contains a suspension of beads. These beads settle very quickly. It is vital that they are mixed immediately before use.
We recommend using the Library Beads (LIB) for most sequencing experiments. However, the Library Solution (LIS) is available for more viscous libraries.
In a new 1.5 ml Eppendorf DNA LoBind tube, prepare the library for loading as follows:
| Reagent | Volume per flow cell |
|---|---|
| Sequencing Buffer (SB) | 100 µl |
| Library Beads (LIB) thoroughly mixed before use, or Library Solution (LIS) | 68 µl |
| DNA library | 32 µl |
| Total | 200 µl |
Note: Library loading volume has been increased to improve array coverage.
It is vital that the inlet port is closed before removing waste to prevent air from being drawn across the sensor array area, which would lead to a significant loss of sequencing channels.
Remove the waste buffer, as follows:
- Ensure the inlet port is closed.
- Insert a P1000 pipette into a waste port and remove the waste buffer
Note: As the inlet port is closed, no fluid should leave the sensor array area.
Slide the inlet port cover clockwise to open the inlet port.

Take care when drawing back buffer from the flow cell. Do not remove more than 20-30 µl, and make sure that the array of pores are covered by buffer at all times. Introducing air bubbles into the array can irreversibly damage pores.
After opening the inlet port, draw back a small volume to remove any air bubbles:
- Set a P1000 pipette tip to 200 µl.
- Insert the tip into the inlet port.
- Turn the wheel until the dial shows 220-230 µl, or until you see a small volume of buffer entering the pipette tip.

Slowly load 500 µl of the priming mix into the inlet port, as follows:
- Using a P1000 pipette, take 500 µl of the priming mix
- Insert the pipette tip into the priming port, ensuring there are no bubbles in the tip
- Slowly twist the pipette wheel down to load the flow cell (if possible with your pipette) or push down the plunger very slowly, leaving a small volume of buffer in the pipette tip.

It is vital that the inlet port is closed before removing waste to prevent air from being drawn across the sensor array area, which would lead to a significant loss of sequencing channels.
Remove waste buffer, as follows:
- Close the inlet port.
- Insert a P1000 pipette into a waste port and remove the waste buffer.
Note: As both the inlet port is closed, no fluid should leave the sensor array area.
Slide the inlet port cover clockwise to open the inlet port.

Take care when drawing back buffer from the flow cell. Do not remove more than 20-30 µl, and make sure that the array of pores are covered by buffer at all times. Introducing air bubbles into the array can irreversibly damage pores.
After opening the inlet port, draw back a small volume to remove any air bubbles:
- Set a P1000 pipette tip to 200 µl.
- Insert the tip into the inlet port.
- Turn the wheel until the dial shows 220-230 µl, or until you see a small volume of buffer entering the pipette tip.

Mix the prepared library gently by pipetting up and down just prior to loading.
Load 200 µl of library into the inlet port using a P1000 pipette.

Close the valve to seal the inlet port.
Install the light shield on your flow cell as soon as library has been loaded for optimal sequencing output.
We recommend leaving the light shield on the flow cell when library is loaded, including during any washing and reloading steps. The shield can be removed when the library has been removed from the flow cell.
If the light shield has been removed from the flow cell, install the light shield as follows:
- Align the inlet port cut out of the light shield with the inlet port cover on the flow cell. The leading edge of the light shield should sit above the flow cell ID.
- Firmly press the light shield around the inlet port cover. The inlet port clip will click into place underneath the inlet port cover.


Close the PromethION lid when ready to start a sequencing run on MinKNOW.
Wait a minimum of 10 minutes after loading the flow cells onto the PromethION before initiating any experiments. This will help to increase the sequencing output.
Resume the sequencing run in MinKNOW to continue data acquisition.
After the final duplex library has been loaded and sequenced, the flow cell can be flushed with deionised water and returned to Oxford Nanopore. For further information, see the "Ending the experiment" step.
21. Day 4: Washing and reloading the PromethION Flow Cell with ultra-long DNA library
材料
- Flow Cell Wash Kit XL (EXP-WSH004-XL)
- 冲洗系绳 UL(FTU)
- 测序芯片冲洗液(FCF)
- 上样溶液 UL(LSU)
- 测序缓冲液 UL(SBU)
耗材
- 1.5 ml Eppendorf DNA LoBind 离心管
仪器
- P1000 移液枪和枪头
- P200 移液枪和枪头
- P20 移液枪和枪头
Day 4: Ultra-long DNA experiment flow cell washing and reloading
We recommend reloading your PromethION Flow Cell with a fresh ultra-long DNA library to maintain high output, using the modified method for reloading a viscous library.
For the Ultra-long DNA experiment, up to three libraries prepared using the Ultra-Long DNA Sequencing Kit V14 (SQK-ULK114) can be loaded on the PromethION Flow Cell during a sequencing run. We recommend washing the flow cell when ~20-25% of active pores are remaining, which typically occurs after ~20-24 hours of sequencing. Washing removes most of the initial library as well as unblocking pores to prepare the flow cell for loading a new library for further sequencing.
Navigate to the Pore Activity or the Pore Scan Results plot to see pore availability. Below is an example of pore states observed on a flow cell before and after wash steps are performed. The red asterisks indicates the reloads. 
Due to the viscosity of the library, the flow cell washing and reloading steps have been modified. It is also recommended to remove the waste fluid before washing the flow cell and before reloading of an ultra-long DNA library after each priming step.
We recommend washing and reloading on days 4 and 5 of this protocol.
We recommend keeping the light shield on the flow cell during washing if a second library will be loaded straight away.
If the flow cell is to be washed and stored, the light shield can be removed.
Place the tube of Wash Mix (WMX) on ice. Do not vortex the tube.
Thaw one tube of Wash Diluent (DIL) at room temperature.
Mix the contents of Wash Diluent (DIL) thoroughly by vortexing, then spin down briefly and place on ice.
In a fresh 1.5 ml Eppendorf DNA LoBind tube, prepare the following Flow Cell Wash Mix:
| Reagent | Volume per flow cell |
|---|---|
| Wash Mix (WMX) | 2 μl |
| Wash Diluent (DIL) | 398 μl |
| Total | 400 μl |
Mix well by pipetting, and place on ice. Do not vortex the tube.
Pause the sequencing experiment in MinKNOW, and leave the flow cell in the device.
Ensure the inlet port is closed and remove the buffer from the waste port, using a P1000 pipette.
The waste fluid can be aspirated from either one of the ports, labelled 2 and 3 on the flow cell.

Slide the inlet port cover clockwise to open the inlet port.

After opening the inlet port, check for a small air bubble under the cover. Draw back a small volume to remove any bubbles:
- Set a P1000 pipette to 200 µl
- Insert the tip into the inlet port
- Turn the wheel until the dial shows 220-230 µl, or until you can see a small volume of buffer entering the pipette tip.

Load 400 µl of the prepared Flow Cell Wash Mix into the flow cell via the inlet port, avoiding the introduction of air.
It may be necessary to use a laboratory wipe to mop up any excess fluid that escapes from the flow cell waste ports 2 and 3.
Close the inlet port and wait for 1 hour.
Ensure the inlet port is closed and remove buffer from the waste port a second time.
The waste fluid can be aspirated from either one of the ports, labelled 2 and 3 on the flow cell.

The buffers used in this process are incompatible with conducting a Flow Cell Check step prior to loading the subsequent library. However, number of available pores will be reported after the next pore scan.
Thaw the Sequencing Buffer UL (SBU), Loading Solution UL (LSU), Flush Tether UL (FTU) and one tube of Flow Cell Flush (FCF) at room temperature and mix by vortexing. Then spin down and place on ice.
In a fresh 1.5 ml Eppendorf DNA LoBind tube, prepare the DNA library for loading as follows using a wide-bore pipette tip for the addition of the DNA library:
| Reagent | Volume per flow cell |
|---|---|
| Sequencing Buffer UL (SBU) | 100 µl |
| Loading Solution UL (LSU) | 10 µl |
| DNA library | 90 µl |
| Total | 200 µl |
Note: ensure the Sequencing Buffer UL (SBU) and Loading Solution UL (LSU) are thoroughly mixed by pipetting before the addition of the DNA library.
Gently mix the prepared DNA library by slowly pipetting ten times using a wide-bore pipette tip.
Incubate at room temperature for 30 minutes then gently mix by slowly pipetting with a wide-bore tip. Visually inspect to ensure the sample is homogenous.
Prepare the flow cell priming mix in a 1.5 ml Eppendorf DNA LoBind tube and mix by vortexing at room temperature.
| Reagent | Volume |
|---|---|
| Flush Tether UL (FTU) | 30 µl |
| Flow Cell Flush (FCF) | 1170 µl |
| Total | 1200 µl |
Slide the inlet port cover clockwise to open.

Take care when drawing back buffer from the flow cell. Do not remove more than 20-30 µl, and make sure that the array of pores are covered by buffer at all times. Introducing air bubbles into the array can irreversibly damage pores.
After opening the inlet port, draw back a small volume to remove any air bubbles:
- Set a P1000 pipette tip to 200 µl.
- Insert the tip into the inlet port.
- Turn the wheel until the dial shows 220-230 µl, or until you see a small volume of buffer entering the pipette tip.

It is vital to wait five minutes between the priming mix flushes to ensure effective removal of the nuclease.
Load 500 µl of the priming mix into the flow cell via the inlet port, avoiding the introduction of air bubbles. Wait five minutes.

Turn the valve to close the inlet port and use a P1000 to remove all fluid from the waste channel through one of the waste ports.
The waste liquid can be aspirated from either one of the ports, labelled 2 and 3.
Slide open the inlet port and load 500 µl of the priming mix into the flow cell via the inlet port to complete a second flow cell flush, avoiding the introduction of air bubbles.

Close the inlet port and use a P1000 to remove all fluid from the waste channel through a waste port again.
Open the inlet port cover of the flow cell in preparation for loading.

Aspirate the DNA library with a wide-bore pipette tip. Ensure there are no air bubbles in the pipette tip. Place the wide-bore pipette tip directly on the inlet port. Slowly depress the pipette to dispense the library into the inlet port.
There can be a delay between depressing the pipette and the library dispensing from the pipette tip. Dispense the library slowly, allowing the library to dispense from the pipette tip before depressing the pipette further.
It is important to dispense the library slowly to prevent air being introduced onto the flow cell.
Note: The DNA library loaded in this step is viscous and may not readily flow through the inlet port into the flow cell. In this case, we recommend applying negative pressure in the flow cell as explained in the steps below.

Using a P200 pipette, set the pipette to 50 µl and insert the tip into Port 2.

Very slowly turn the wheel of the pipette to pull the DNA library into the inlet port. Closely watch the DNA library on the inlet port and completely remove the pipette as soon as the library starts to be pulled into the port.
This step is required if the DNA library has not been fully absorbed into the inlet port.
Note: Take care to not apply too much negative pressure too quickly to avoid bringing air bubbles into the flow cell. Air bubbles will cause irreversible damage to the flow cell.

Close the valve to seal the inlet port.
Install the light shield on your flow cell as soon as library has been loaded for optimal sequencing output.
We recommend leaving the light shield on the flow cell when library is loaded, including during any washing and reloading steps. The shield can be removed when the library has been removed from the flow cell.
If the light shield has been removed from the flow cell, install the light shield as follows:
- Align the inlet port cut out of the light shield with the inlet port cover on the flow cell. The leading edge of the light shield should sit above the flow cell ID.
- Firmly press the light shield around the inlet port cover. The inlet port clip will click into place underneath the inlet port cover.


Resume the sequencing run on MinKNOW to continue data acquisition.
22. Day 5: Washing and reloading Pore-C library on the PromethION Flow Cell
材料
- Flow Cell Wash Kit XL (EXP-WSH004-XL)
- 测序辅助扩展包 V14(EXP-AUX003)
- 测序缓冲液(SB)
- 文库溶液(LIS)
- 文库颗粒(LIB)
- 测序芯片系绳(FCT)
- 测序芯片冲洗液(FCF)
耗材
- 1.5 ml Eppendorf DNA LoBind 离心管
仪器
- P1000 移液枪和枪头
- P20 移液枪和枪头
- 盛有冰的冰桶
Day 5: Pore-C experiment flow cell washing and reloading
We recommend washing and reloading your PromethION Flow Cell with a new library to maintain high data acquisition.
For the Pore-C experiment, up to four libraries prepared using the Pore-C protocol and Ligation Sequencing Kit XL V14 (SQK-LSK114-XL) can be loaded on the PromethION Flow Cell during a sequencing run. We recommend washing your flow cell when ~20-25% of active pores are remaining, which typically occurs after ~18 hours of sequencing. Washing removes most of the initial library as well as unblocking pores to prepare a flow cell for loading a fresh library for further sequencing. Pore-C DNA extracts are prone to blocking and may require closer monitoring and washing earlier into the sequencing run than the duplex experiment to optimise sequencing output.
Navigate to the Pore Activity or the Pore Scan Results plot to see pore availability. Below is an example of pore states observed on a flow cell before and after wash steps are performed. The red asterisks indicates the reloads. 
We recommend reloading your Pore-C experiment on days 5, 6 and 7.
Washing and reloading a PromethION Flow Cell video
This video will show you how to wash a flow cell after a sequencing run and how to load a new library.
We recommend keeping the light shield on the flow cell during washing if a second library will be loaded straight away.
If the flow cell is to be washed and stored, the light shield can be removed.
Place the tube of Wash Mix (WMX) on ice. Do not vortex the tube.
Thaw one tube of Wash Diluent (DIL) at room temperature.
Mix the contents of Wash Diluent (DIL) thoroughly by vortexing, then spin down briefly and place on ice.
In a fresh 1.5 ml Eppendorf DNA LoBind tube, prepare the following Flow Cell Wash Mix:
| Reagent | Volume per flow cell |
|---|---|
| Wash Mix (WMX) | 2 μl |
| Wash Diluent (DIL) | 398 μl |
| Total | 400 μl |
Mix well by pipetting, and place on ice. Do not vortex the tube.
Pause the sequencing experiment in MinKNOW, and leave the flow cell in the device.
It is vital that the inlet port is closed before removing waste to prevent air from being drawn across the sensor array area, which would lead to a significant loss of sequencing channels.
Remove waste buffer, as follows:
- Close the inlet port.
- Insert a P1000 pipette into a waste port and remove the waste buffer.
Note: As both the inlet port is closed, no fluid should leave the sensor array area.
Slide the inlet port cover clockwise to open the inlet port.

Take care when drawing back buffer from the flow cell. Do not remove more than 20-30 µl, and make sure that the array of pores are covered by buffer at all times. Introducing air bubbles into the array can irreversibly damage pores.
After opening the inlet port, check for a small air bubble under the cover. Draw back a small volume to remove any bubbles:
- Set a P1000 pipette to 200 µl
- Insert the tip into the inlet port
- Turn the wheel until the dial shows 220-230 µl, or until you can see a small volume of buffer entering the pipette tip.

Slowly load 200 µl of the prepared flow cell wash mix into the inlet port, as follows:
- Using a P1000 pipette, take 200 µl of the flow cell wash mix
- Insert the pipette tip into the inlet port, ensuring there are no bubbles in the tip
- Slowly twist the pipette wheel down to load the flow cell (if possible with your pipette) or push down the plunger very slowly, leaving a small volume of buffer in the pipette tip.
- Set a timer for a 5 minute incubation.
Once the 5 minute incubation time is complete, carefully load the remaining 200 µl of the prepared flow cell wash mix into the inlet port, as follows:
- Using a P1000 pipette, take 200 µl of the flow cell wash mix
- Insert the pipette tip into the inlet port, ensuring there are no bubbles in the tip
- Slowly twist the pipette wheel down to load the flow cell (if possible with your pipette) or push down the plunger very slowly, leaving a small volume of buffer in the pipette tip.
Close the inlet port and wait for 1 hour.
It is vital that the inlet port is closed before removing waste to prevent air from being drawn across the sensor array area, which would lead to a significant loss of sequencing channels.
Remove the waste buffer, as follows:
- Ensure the inlet port is closed.
- Insert a P1000 pipette into a waste port and remove the waste buffer
Note: As the inlet port is closed, no fluid should leave the sensor array area.
The buffers used in this process are incompatible with conducting a Flow Cell Check step prior to loading the subsequent library. However, number of available pores will be reported after the next pore scan.
Thaw the Sequencing Buffer (SB), Library Beads (LIB) or Library Solution (LIS, if using), Flow Cell Tether (FCT) and Flow Cell Flush (FCF) at room temperature before mixing by vortexing. Then spin down and store on ice.
Prepare the flow cell priming mix in a suitable tube for the number of flow cells to flush. Once combined, mix well by briefly vortexing.
| Reagents | Volume per flow cell |
|---|---|
| Flow Cell Flush (FCF) | 1,170 µl |
| Flow Cell Tether (FCT) | 30 µl |
| Total volume | 1,200 µl |
Slide the inlet port cover clockwise to open.

Take care when drawing back buffer from the flow cell. Do not remove more than 20-30 µl, and make sure that the array of pores are covered by buffer at all times. Introducing air bubbles into the array can irreversibly damage pores.
After opening the inlet port, draw back a small volume to remove any air bubbles:
- Set a P1000 pipette tip to 200 µl.
- Insert the tip into the inlet port.
- Turn the wheel until the dial shows 220-230 µl, or until you see a small volume of buffer entering the pipette tip.

Slowly load 500 µl of the priming mix into the inlet port, as follows:
- Using a P1000 pipette, take 500 µl of the priming mix
- Insert the pipette tip into the priming port, ensuring there are no bubbles in the tip
- Slowly twist the pipette wheel down to load the flow cell (if possible with your pipette) or push down the plunger very slowly, leaving a small volume of buffer in the pipette tip.

It is vital to wait five minutes between the priming mix flushes to ensure effective removal of the nuclease.
Close the inlet port and wait five minutes.
During this time, prepare the library for loading using the next steps in the protocol.
Thoroughly mix the contents of the Library Beads (LIB) by pipetting.
The Library Beads (LIB) tube contains a suspension of beads. These beads settle very quickly. It is vital that they are mixed immediately before use.
We recommend using the Library Beads (LIB) for most sequencing experiments. However, the Library Solution (LIS) is available for more viscous libraries.
In a new 1.5 ml Eppendorf DNA LoBind tube, prepare the library for loading as follows:
| Reagent | Volume per flow cell |
|---|---|
| Sequencing Buffer (SB) | 100 µl |
| Library Beads (LIB) thoroughly mixed before use, or Library Solution (LIS) | 68 µl |
| DNA library | 32 µl |
| Total | 200 µl |
Note: Library loading volume has been increased to improve array coverage.
It is vital that the inlet port is closed before removing waste to prevent air from being drawn across the sensor array area, which would lead to a significant loss of sequencing channels.
Remove the waste buffer, as follows:
- Ensure the inlet port is closed.
- Insert a P1000 pipette into a waste port and remove the waste buffer
Note: As the inlet port is closed, no fluid should leave the sensor array area.
Slide the inlet port cover clockwise to open the inlet port.

Take care when drawing back buffer from the flow cell. Do not remove more than 20-30 µl, and make sure that the array of pores are covered by buffer at all times. Introducing air bubbles into the array can irreversibly damage pores.
After opening the inlet port, draw back a small volume to remove any air bubbles:
- Set a P1000 pipette tip to 200 µl.
- Insert the tip into the inlet port.
- Turn the wheel until the dial shows 220-230 µl, or until you see a small volume of buffer entering the pipette tip.

Slowly load 500 µl of the priming mix into the inlet port, as follows:
- Using a P1000 pipette, take 500 µl of the priming mix
- Insert the pipette tip into the priming port, ensuring there are no bubbles in the tip
- Slowly twist the pipette wheel down to load the flow cell (if possible with your pipette) or push down the plunger very slowly, leaving a small volume of buffer in the pipette tip.

It is vital that the inlet port is closed before removing waste to prevent air from being drawn across the sensor array area, which would lead to a significant loss of sequencing channels.
Remove waste buffer, as follows:
- Close the inlet port.
- Insert a P1000 pipette into a waste port and remove the waste buffer.
Note: As both the inlet port is closed, no fluid should leave the sensor array area.
Slide the inlet port cover clockwise to open.

Take care when drawing back buffer from the flow cell. Do not remove more than 20-30 µl, and make sure that the array of pores are covered by buffer at all times. Introducing air bubbles into the array can irreversibly damage pores.
After opening the inlet port, draw back a small volume to remove any air bubbles:
- Set a P1000 pipette tip to 200 µl.
- Insert the tip into the inlet port.
- Turn the wheel until the dial shows 220-230 µl, or until you see a small volume of buffer entering the pipette tip.

Mix the prepared library gently by pipetting up and down just prior to loading.
Load 200 µl of library into the inlet port using a P1000 pipette.

Close the valve to seal the inlet port.
Install the light shield on your flow cell as soon as library has been loaded for optimal sequencing output.
We recommend leaving the light shield on the flow cell when library is loaded, including during any washing and reloading steps. The shield can be removed when the library has been removed from the flow cell.
If the light shield has been removed from the flow cell, install the light shield as follows:
- Align the inlet port cut out of the light shield with the inlet port cover on the flow cell. The leading edge of the light shield should sit above the flow cell ID.
- Firmly press the light shield around the inlet port cover. The inlet port clip will click into place underneath the inlet port cover.


Close the PromethION lid when ready to start a sequencing run on MinKNOW.
Wait a minimum of 10 minutes after loading the flow cells onto the PromethION before initiating any experiments. This will help to increase the sequencing output.
Resume the sequencing run in MinKNOW to continue data acquisition.
23. Day 5: Washing and reloading the PromethION Flow Cell with ultra-long DNA library
材料
- Flow Cell Wash Kit XL (EXP-WSH004-XL)
- 冲洗系绳 UL(FTU)
- 测序芯片冲洗液(FCF)
- 上样溶液 UL(LSU)
- 测序缓冲液 UL(SBU)
耗材
- 1.5 ml Eppendorf DNA LoBind 离心管
仪器
- P1000 移液枪和枪头
- P200 移液枪和枪头
- P20 移液枪和枪头
Day 5: Ultra-long DNA experiment flow cell washing and reloading
We recommend reloading your PromethION Flow Cell with a fresh ultra-long DNA library to maintain high output, using the modified method for reloading a viscous library.
For the Ultra-long DNA experiment, up to three libraries prepared using the Ultra-Long DNA Sequencing Kit V14 (SQK-ULK114) can be loaded on the PromethION flow cell during a sequencing run. We recommend washing the flow cell when ~20-25% of active pores are remaining, which typically occurs after ~20-24 hours of sequencing. Washing removes most of the initial library as well as unblocking pores to prepare the flow cell for loading a new library for further sequencing.
Navigate to the Pore Activity or the Pore Scan Results plot to see pore availability. Below is an example of pore states observed on a flow cell before and after wash steps are performed. The red asterisks indicates the reloads. 
Due to the viscosity of the library, the flow cell washing and reloading steps have been modified. It is also recommended to remove the waste fluid before washing the flow cell and before reloading an ultra-long DNA library after each priming step.
We recommend washing and reloading on days 4 and 5 of this protocol.
Washing and reloading a PromethION Flow Cell video
This video will show you how to wash a flow cell after a sequencing run and how to load a new library.
We recommend keeping the light shield on the flow cell during washing if a second library will be loaded straight away.
If the flow cell is to be washed and stored, the light shield can be removed.
Place the tube of Wash Mix (WMX) on ice. Do not vortex the tube.
Thaw one tube of Wash Diluent (DIL) at room temperature.
Mix the contents of Wash Diluent (DIL) thoroughly by vortexing, then spin down briefly and place on ice.
In a fresh 1.5 ml Eppendorf DNA LoBind tube, prepare the following Flow Cell Wash Mix:
| Reagent | Volume per flow cell |
|---|---|
| Wash Mix (WMX) | 2 μl |
| Wash Diluent (DIL) | 398 μl |
| Total | 400 μl |
Mix well by pipetting, and place on ice. Do not vortex the tube.
Pause the sequencing experiment in MinKNOW, and leave the flow cell in the device.
It is vital that the inlet port is closed before removing waste to prevent air from being drawn across the sensor array area, which would lead to a significant loss of sequencing channels.
Remove waste buffer, as follows:
- Close the inlet port.
- Insert a P1000 pipette into a waste port and remove the waste buffer.
Note: As both the inlet port is closed, no fluid should leave the sensor array area.
Slide the inlet port cover clockwise to open the inlet port.

Take care when drawing back buffer from the flow cell. Do not remove more than 20-30 µl, and make sure that the array of pores are covered by buffer at all times. Introducing air bubbles into the array can irreversibly damage pores.
After opening the inlet port, check for a small air bubble under the cover. Draw back a small volume to remove any bubbles:
- Set a P1000 pipette to 200 µl
- Insert the tip into the inlet port
- Turn the wheel until the dial shows 220-230 µl, or until you can see a small volume of buffer entering the pipette tip.

Slowly load 200 µl of the prepared flow cell wash mix into the inlet port, as follows:
- Using a P1000 pipette, take 200 µl of the flow cell wash mix
- Insert the pipette tip into the inlet port, ensuring there are no bubbles in the tip
- Slowly twist the pipette wheel down to load the flow cell (if possible with your pipette) or push down the plunger very slowly, leaving a small volume of buffer in the pipette tip.
- Set a timer for a 5 minute incubation.
Once the 5 minute incubation time is complete, carefully load the remaining 200 µl of the prepared flow cell wash mix into the inlet port, as follows:
- Using a P1000 pipette, take 200 µl of the flow cell wash mix
- Insert the pipette tip into the inlet port, ensuring there are no bubbles in the tip
- Slowly twist the pipette wheel down to load the flow cell (if possible with your pipette) or push down the plunger very slowly, leaving a small volume of buffer in the pipette tip.
Close the inlet port and wait for 1 hour.
It is vital that the inlet port is closed before removing waste to prevent air from being drawn across the sensor array area, which would lead to a significant loss of sequencing channels.
Remove waste buffer, as follows:
- Close the inlet port.
- Insert a P1000 pipette into a waste port and remove the waste buffer.
Note: As both the inlet port is closed, no fluid should leave the sensor array area.
The buffers used in this process are incompatible with conducting a Flow Cell Check step prior to loading the subsequent library. However, number of available pores will be reported after the next pore scan.
Thaw the Sequencing Buffer UL (SBU), Loading Solution UL (LSU), Flush Tether UL (FTU) and one tube of Flow Cell Flush (FCF) at room temperature and mix by vortexing. Then spin down and place on ice.
In a fresh 1.5 ml Eppendorf DNA LoBind tube, prepare the DNA library for loading as follows using a wide-bore pipette tip for the addition of the DNA library:
| Reagent | Volume per flow cell |
|---|---|
| Sequencing Buffer UL (SBU) | 100 µl |
| Loading Solution UL (LSU) | 10 µl |
| DNA library | 90 µl |
| Total | 200 µl |
Note: ensure the Sequencing Buffer UL (SBU) and Loading Solution UL (LSU) are thoroughly mixed by pipetting before the addition of the DNA library.
Gently mix the prepared DNA library by slowly pipetting ten times using a wide-bore pipette tip.
Incubate at room temperature for 30 minutes then gently mix by slowly pipetting with a wide-bore tip. Visually inspect to ensure the sample is homogenous.
Prepare the flow cell priming mix in a 1.5 ml Eppendorf DNA LoBind tube and mix by vortexing at room temperature.
| Reagent | Volume |
|---|---|
| Flush Tether UL (FTU) | 30 µl |
| Flow Cell Flush (FCF) | 1170 µl |
| Total | 1200 µl |
Slide the inlet port cover clockwise to open.

Take care when drawing back buffer from the flow cell. Do not remove more than 20-30 µl, and make sure that the array of pores are covered by buffer at all times. Introducing air bubbles into the array can irreversibly damage pores.
After opening the inlet port, draw back a small volume to remove any air bubbles:
- Set a P1000 pipette tip to 200 µl.
- Insert the tip into the inlet port.
- Turn the wheel until the dial shows 220-230 µl, or until you see a small volume of buffer entering the pipette tip.

Slowly load 500 µl of the priming mix into the inlet port, as follows:
- Using a P1000 pipette, take 500 µl of the priming mix
- Insert the pipette tip into the priming port, ensuring there are no bubbles in the tip
- Slowly twist the pipette wheel down to load the flow cell (if possible with your pipette) or push down the plunger very slowly, leaving a small volume of buffer in the pipette tip.

It is vital to wait five minutes between the priming mix flushes to ensure effective removal of the nuclease.
Close the inlet port and wait five minutes.
During this time, prepare the library for loading using the next steps in the protocol.
Turn the valve to close the inlet port and use a P1000 to remove all fluid from the waste channel through one of the waste ports.
The waste liquid can be aspirated from either one of the ports, labelled 2 and 3.
Slide open the inlet port and load 500 µl of the priming mix into the flow cell via the inlet port to complete a second flow cell flush, avoiding the introduction of air bubbles.

Close the inlet port and use a P1000 to remove all fluid from the waste channel through a waste port again.
Open the inlet port cover of the flow cell in preparation for loading.

Aspirate the DNA library with a wide-bore pipette tip. Ensure there are no air bubbles in the pipette tip. Place the wide-bore pipette tip directly on the inlet port. Slowly depress the pipette to dispense the library into the inlet port.
There can be a delay between depressing the pipette and the library dispensing from the pipette tip. Dispense the library slowly, allowing the library to dispense from the pipette tip before depressing the pipette further.
It is important to dispense the library slowly to prevent air being introduced onto the flow cell.
Note: The DNA library loaded in this step is viscous and may not readily flow through the inlet port into the flow cell. In this case, we recommend applying negative pressure in the flow cell as explained in the steps below.

Using a P200 pipette, set the pipette to 50 µl and insert the tip into Port 2.

Very slowly turn the wheel of the pipette to pull the DNA library into the inlet port. Closely watch the DNA library on the inlet port and completely remove the pipette as soon as the library starts to be pulled into the port.
This step is required if the DNA library has not been fully absorbed into the inlet port.
Note: Take care to not apply too much negative pressure too quickly to avoid bringing air bubbles into the flow cell. Air bubbles will cause irreversible damage to the flow cell.

Close the valve to seal the inlet port.
Install the light shield on your flow cell as soon as library has been loaded for optimal sequencing output.
We recommend leaving the light shield on the flow cell when library is loaded, including during any washing and reloading steps. The shield can be removed when the library has been removed from the flow cell.
If the light shield has been removed from the flow cell, install the light shield as follows:
- Align the inlet port cut out of the light shield with the inlet port cover on the flow cell. The leading edge of the light shield should sit above the flow cell ID.
- Firmly press the light shield around the inlet port cover. The inlet port clip will click into place underneath the inlet port cover.


Resume the sequencing run in MinKNOW to continue data acquisition.
After the final ultra-long DNA library has been loaded and sequenced, the flow cell can be flushed with deionised water and returned to Oxford Nanopore. For further information, see the "Ending the experiment" step.
24. Day 6: Washing and reloading Pore-C library on the PromethION Flow Cell
材料
- Flow Cell Wash Kit XL (EXP-WSH004-XL)
- 测序辅助扩展包 V14(EXP-AUX003)
- 测序缓冲液(SB)
- 文库溶液(LIS)
- 文库颗粒(LIB)
- 测序芯片冲洗液(FCF)
- 测序芯片系绳(FCT)
仪器
- P1000 移液枪和枪头
- P20 移液枪和枪头
- 盛有冰的冰桶
Day 6: Pore-C experiment flow cell washing and reloading
We recommend washing and reloading your PromethION Flow Cell with a new library to maintain high data acquisition.
For the Pore-C experiment, up to four libraries prepared using the Pore-C protocol and Ligation Sequencing Kit XL V14 (SQK-LSK114-XL) can be loaded on the PromethION Flow Cell during a sequencing run. We recommend washing your flow cell when ~20-25% of active pores are remaining, which typical occurs after ~18 hours of sequencing. Washing removes most of the initial library as well as unblocking pores to prepare a flow cell for loading a fresh library for further sequencing. Pore-C DNA extracts are prone to blocking and may require closer monitoring and washing earlier into the sequencing run than the duplex experiment to optimise sequencing output.
Navigate to the Pore Activity or the Pore Scan Results plot to see pore availability. Below is an example of pore states observed on a flow cell before and after wash steps are performed. The red asterisks indicates the reloads. 
We recommend reloading your Pore-C experiment on days 5, 6 and 7.
Washing and reloading a PromethION Flow Cell video
This video will show you how to wash a flow cell after a sequencing run and how to load a new library.
We recommend keeping the light shield on the flow cell during washing if a second library will be loaded straight away.
If the flow cell is to be washed and stored, the light shield can be removed.
Place the tube of Wash Mix (WMX) on ice. Do not vortex the tube.
Thaw one tube of Wash Diluent (DIL) at room temperature.
Mix the contents of Wash Diluent (DIL) thoroughly by vortexing, then spin down briefly and place on ice.
In a fresh 1.5 ml Eppendorf DNA LoBind tube, prepare the following Flow Cell Wash Mix:
| Reagent | Volume per flow cell |
|---|---|
| Wash Mix (WMX) | 2 μl |
| Wash Diluent (DIL) | 398 μl |
| Total | 400 μl |
Mix well by pipetting, and place on ice. Do not vortex the tube.
Pause the sequencing experiment in MinKNOW, and leave the flow cell in the device.
It is vital that the inlet port is closed before removing waste to prevent air from being drawn across the sensor array area, which would lead to a significant loss of sequencing channels.
Remove waste buffer, as follows:
- Close the inlet port.
- Insert a P1000 pipette into a waste port and remove the waste buffer.
Note: As both the inlet port is closed, no fluid should leave the sensor array area.
Slide the inlet port cover clockwise to open the inlet port.

Take care when drawing back buffer from the flow cell. Do not remove more than 20-30 µl, and make sure that the array of pores are covered by buffer at all times. Introducing air bubbles into the array can irreversibly damage pores.
After opening the inlet port, check for a small air bubble under the cover. Draw back a small volume to remove any bubbles:
- Set a P1000 pipette to 200 µl
- Insert the tip into the inlet port
- Turn the wheel until the dial shows 220-230 µl, or until you can see a small volume of buffer entering the pipette tip.

Slowly load 200 µl of the prepared flow cell wash mix into the inlet port, as follows:
- Using a P1000 pipette, take 200 µl of the flow cell wash mix
- Insert the pipette tip into the inlet port, ensuring there are no bubbles in the tip
- Slowly twist the pipette wheel down to load the flow cell (if possible with your pipette) or push down the plunger very slowly, leaving a small volume of buffer in the pipette tip.
- Set a timer for a 5 minute incubation.
Once the 5 minute incubation time is complete, carefully load the remaining 200 µl of the prepared flow cell wash mix into the inlet port, as follows:
- Using a P1000 pipette, take 200 µl of the flow cell wash mix
- Insert the pipette tip into the inlet port, ensuring there are no bubbles in the tip
- Slowly twist the pipette wheel down to load the flow cell (if possible with your pipette) or push down the plunger very slowly, leaving a small volume of buffer in the pipette tip.
Close the inlet port and wait for 1 hour.
It is vital that the inlet port is closed before removing waste to prevent air from being drawn across the sensor array area, which would lead to a significant loss of sequencing channels.
Remove the waste buffer, as follows:
- Ensure the inlet port is closed.
- Insert a P1000 pipette into a waste port and remove the waste buffer
Note: As the inlet port is closed, no fluid should leave the sensor array area.
The buffers used in this process are incompatible with conducting a Flow Cell Check step prior to loading the subsequent library. However, number of available pores will be reported after the next pore scan.
Thaw the Sequencing Buffer (SB), Library Beads (LIB) or Library Solution (LIS, if using), Flow Cell Tether (FCT) and Flow Cell Flush (FCF) at room temperature before mixing by vortexing. Then spin down and store on ice.
Prepare the flow cell priming mix in a suitable tube for the number of flow cells to flush. Once combined, mix well by briefly vortexing.
| Reagents | Volume per flow cell |
|---|---|
| Flow Cell Flush (FCF) | 1,170 µl |
| Flow Cell Tether (FCT) | 30 µl |
| Total volume | 1,200 µl |
Slide the inlet port cover clockwise to open.

Take care when drawing back buffer from the flow cell. Do not remove more than 20-30 µl, and make sure that the array of pores are covered by buffer at all times. Introducing air bubbles into the array can irreversibly damage pores.
After opening the inlet port, draw back a small volume to remove any air bubbles:
- Set a P1000 pipette tip to 200 µl.
- Insert the tip into the inlet port.
- Turn the wheel until the dial shows 220-230 µl, or until you see a small volume of buffer entering the pipette tip.

Slowly load 500 µl of the priming mix into the inlet port, as follows:
- Using a P1000 pipette, take 500 µl of the priming mix
- Insert the pipette tip into the priming port, ensuring there are no bubbles in the tip
- Slowly twist the pipette wheel down to load the flow cell (if possible with your pipette) or push down the plunger very slowly, leaving a small volume of buffer in the pipette tip.

It is vital to wait five minutes between the priming mix flushes to ensure effective removal of the nuclease.
Close the inlet port and wait five minutes.
During this time, prepare the library for loading using the next steps in the protocol.
Thoroughly mix the contents of the Library Beads (LIB) by pipetting.
The Library Beads (LIB) tube contains a suspension of beads. These beads settle very quickly. It is vital that they are mixed immediately before use.
We recommend using the Library Beads (LIB) for most sequencing experiments. However, the Library Solution (LIS) is available for more viscous libraries.
In a new 1.5 ml Eppendorf DNA LoBind tube, prepare the library for loading as follows:
| Reagent | Volume per flow cell |
|---|---|
| Sequencing Buffer (SB) | 100 µl |
| Library Beads (LIB) thoroughly mixed before use, or Library Solution (LIS) | 68 µl |
| DNA library | 32 µl |
| Total | 200 µl |
Note: Library loading volume has been increased to improve array coverage.
It is vital that the inlet port is closed before removing waste to prevent air from being drawn across the sensor array area, which would lead to a significant loss of sequencing channels.
Remove the waste buffer, as follows:
- Ensure the inlet port is closed.
- Insert a P1000 pipette into a waste port and remove the waste buffer
Note: As the inlet port is closed, no fluid should leave the sensor array area.
Slide the inlet port cover clockwise to open.

Take care when drawing back buffer from the flow cell. Do not remove more than 20-30 µl, and make sure that the array of pores are covered by buffer at all times. Introducing air bubbles into the array can irreversibly damage pores.
After opening the inlet port, draw back a small volume to remove any air bubbles:
- Set a P1000 pipette tip to 200 µl.
- Insert the tip into the inlet port.
- Turn the wheel until the dial shows 220-230 µl, or until you see a small volume of buffer entering the pipette tip.

Slowly load 500 µl of the priming mix into the inlet port, as follows:
- Using a P1000 pipette, take 500 µl of the priming mix
- Insert the pipette tip into the priming port, ensuring there are no bubbles in the tip
- Slowly twist the pipette wheel down to load the flow cell (if possible with your pipette) or push down the plunger very slowly, leaving a small volume of buffer in the pipette tip.

It is vital that the inlet port is closed before removing waste to prevent air from being drawn across the sensor array area, which would lead to a significant loss of sequencing channels.
Remove waste buffer, as follows:
- Close the inlet port.
- Insert a P1000 pipette into a waste port and remove the waste buffer.
Note: As both the inlet port is closed, no fluid should leave the sensor array area.
Slide the inlet port cover clockwise to open.

Take care when drawing back buffer from the flow cell. Do not remove more than 20-30 µl, and make sure that the array of pores are covered by buffer at all times. Introducing air bubbles into the array can irreversibly damage pores.
After opening the inlet port, draw back a small volume to remove any air bubbles:
- Set a P1000 pipette tip to 200 µl.
- Insert the tip into the inlet port.
- Turn the wheel until the dial shows 220-230 µl, or until you see a small volume of buffer entering the pipette tip.

Mix the prepared library gently by pipetting up and down just prior to loading.
Load 200 µl of library into the inlet port using a P1000 pipette.

Close the valve to seal the inlet port.
Install the light shield on your flow cell as soon as library has been loaded for optimal sequencing output.
We recommend leaving the light shield on the flow cell when library is loaded, including during any washing and reloading steps. The shield can be removed when the library has been removed from the flow cell.
If the light shield has been removed from the flow cell, install the light shield as follows:
- Align the inlet port cut out of the light shield with the inlet port cover on the flow cell. The leading edge of the light shield should sit above the flow cell ID.
- Firmly press the light shield around the inlet port cover. The inlet port clip will click into place underneath the inlet port cover.


Close the PromethION lid when ready to start a sequencing run on MinKNOW.
Wait a minimum of 10 minutes after loading the flow cells onto the PromethION before initiating any experiments. This will help to increase the sequencing output.
Resume the sequencing run in MinKNOW to continue data acquisition.
25. Day 7: Washing and reloading Pore-C library on the PromethION Flow Cell
材料
- Flow Cell Wash Kit XL (EXP-WSH004-XL)
- 测序辅助扩展包 V14(EXP-AUX003)
- 测序缓冲液(SB)
- 文库颗粒(LIB)
- 文库溶液(LIS)
- 测序芯片系绳(FCT)
- 测序芯片冲洗液(FCF)
耗材
- 1.5 ml Eppendorf DNA LoBind 离心管
仪器
- P1000 移液枪和枪头
- P20 移液枪和枪头
- 盛有冰的冰桶
Day 7: Pore-C experiment flow cell washing and reloading
We recommend washing and reloading your PromethION Flow Cell with a new library to maintain high data acquisition.
For the Pore-C experiment, up to four libraries prepared using the Pore-C protocol and Ligation Sequencing Kit XL V14 (SQK-LSK114-XL) can be loaded on the PromethION flow cell during a sequencing run. We recommend washing your flow cell when ~20-25% of active pores are remaining, which typically occurs after ~18 hours of sequencing. Washing removes most of the initial library as well as unblocking pores to prepare a flow cell for loading a fresh library for further sequencing. Pore-C DNA extracts are prone to blocking and may require closer monitoring and washing earlier into the sequencing run than the duplex experiment to optimise sequencing output.
Navigate to the Pore Activity or the Pore Scan Results plot to see pore availability. Below is an example of pore states observed on a flow cell before and after wash steps are performed. The red asterisks indicates the reloads. 
We recommend reloading your Pore-C experiment on days 5, 6 and 7.
Washing and reloading a PromethION Flow Cell video
This video will show you how to wash a flow cell after a sequencing run and how to load a new library.
We recommend keeping the light shield on the flow cell during washing if a second library will be loaded straight away.
If the flow cell is to be washed and stored, the light shield can be removed.
Place the tube of Wash Mix (WMX) on ice. Do not vortex the tube.
Thaw one tube of Wash Diluent (DIL) at room temperature.
Mix the contents of Wash Diluent (DIL) thoroughly by vortexing, then spin down briefly and place on ice.
In a fresh 1.5 ml Eppendorf DNA LoBind tube, prepare the following Flow Cell Wash Mix:
| Reagent | Volume per flow cell |
|---|---|
| Wash Mix (WMX) | 2 μl |
| Wash Diluent (DIL) | 398 μl |
| Total | 400 μl |
Mix well by pipetting, and place on ice. Do not vortex the tube.
Pause the sequencing experiment in MinKNOW, and leave the flow cell in the device.
It is vital that the inlet port is closed before removing waste to prevent air from being drawn across the sensor array area, which would lead to a significant loss of sequencing channels.
Remove waste buffer, as follows:
- Close the inlet port.
- Insert a P1000 pipette into a waste port and remove the waste buffer.
Note: As both the inlet port is closed, no fluid should leave the sensor array area.
Slide the inlet port cover clockwise to open the inlet port.

Take care when drawing back buffer from the flow cell. Do not remove more than 20-30 µl, and make sure that the array of pores are covered by buffer at all times. Introducing air bubbles into the array can irreversibly damage pores.
After opening the inlet port, check for a small air bubble under the cover. Draw back a small volume to remove any bubbles:
- Set a P1000 pipette to 200 µl
- Insert the tip into the inlet port
- Turn the wheel until the dial shows 220-230 µl, or until you can see a small volume of buffer entering the pipette tip.

Slowly load 200 µl of the prepared flow cell wash mix into the inlet port, as follows:
- Using a P1000 pipette, take 200 µl of the flow cell wash mix
- Insert the pipette tip into the inlet port, ensuring there are no bubbles in the tip
- Slowly twist the pipette wheel down to load the flow cell (if possible with your pipette) or push down the plunger very slowly, leaving a small volume of buffer in the pipette tip.
- Set a timer for a 5 minute incubation.
Once the 5 minute incubation time is complete, carefully load the remaining 200 µl of the prepared flow cell wash mix into the inlet port, as follows:
- Using a P1000 pipette, take 200 µl of the flow cell wash mix
- Insert the pipette tip into the inlet port, ensuring there are no bubbles in the tip
- Slowly twist the pipette wheel down to load the flow cell (if possible with your pipette) or push down the plunger very slowly, leaving a small volume of buffer in the pipette tip.
Close the inlet port and wait for 1 hour.
It is vital that the inlet port is closed before removing waste to prevent air from being drawn across the sensor array area, which would lead to a significant loss of sequencing channels.
Remove the waste buffer, as follows:
- Ensure the inlet port is closed.
- Insert a P1000 pipette into a waste port and remove the waste buffer
Note: As the inlet port is closed, no fluid should leave the sensor array area.
The buffers used in this process are incompatible with conducting a Flow Cell Check step prior to loading the subsequent library. However, number of available pores will be reported after the next pore scan.
Thaw the Sequencing Buffer (SB), Library Beads (LIB) or Library Solution (LIS, if using), Flow Cell Tether (FCT) and Flow Cell Flush (FCF) at room temperature before mixing by vortexing. Then spin down and store on ice.
Prepare the flow cell priming mix in a suitable tube for the number of flow cells to flush. Once combined, mix well by briefly vortexing.
| Reagents | Volume per flow cell |
|---|---|
| Flow Cell Flush (FCF) | 1,170 µl |
| Flow Cell Tether (FCT) | 30 µl |
| Total volume | 1,200 µl |
Slide the inlet port cover clockwise to open.

Take care when drawing back buffer from the flow cell. Do not remove more than 20-30 µl, and make sure that the array of pores are covered by buffer at all times. Introducing air bubbles into the array can irreversibly damage pores.
After opening the inlet port, draw back a small volume to remove any air bubbles:
- Set a P1000 pipette tip to 200 µl.
- Insert the tip into the inlet port.
- Turn the wheel until the dial shows 220-230 µl, or until you see a small volume of buffer entering the pipette tip.

Slowly load 500 µl of the priming mix into the inlet port, as follows:
- Using a P1000 pipette, take 500 µl of the priming mix
- Insert the pipette tip into the priming port, ensuring there are no bubbles in the tip
- Slowly twist the pipette wheel down to load the flow cell (if possible with your pipette) or push down the plunger very slowly, leaving a small volume of buffer in the pipette tip.

It is vital to wait five minutes between the priming mix flushes to ensure effective removal of the nuclease.
Close the inlet port and wait five minutes.
During this time, prepare the library for loading using the next steps in the protocol.
Thoroughly mix the contents of the Library Beads (LIB) by pipetting.
The Library Beads (LIB) tube contains a suspension of beads. These beads settle very quickly. It is vital that they are mixed immediately before use.
We recommend using the Library Beads (LIB) for most sequencing experiments. However, the Library Solution (LIS) is available for more viscous libraries.
In a new 1.5 ml Eppendorf DNA LoBind tube, prepare the library for loading as follows:
| Reagent | Volume per flow cell |
|---|---|
| Sequencing Buffer (SB) | 100 µl |
| Library Beads (LIB) thoroughly mixed before use, or Library Solution (LIS) | 68 µl |
| DNA library | 32 µl |
| Total | 200 µl |
Note: Library loading volume has been increased to improve array coverage.
It is vital that the inlet port is closed before removing waste to prevent air from being drawn across the sensor array area, which would lead to a significant loss of sequencing channels.
Remove the waste buffer, as follows:
- Ensure the inlet port is closed.
- Insert a P1000 pipette into a waste port and remove the waste buffer
Note: As the inlet port is closed, no fluid should leave the sensor array area.
Slide the inlet port cover clockwise to open.

Take care when drawing back buffer from the flow cell. Do not remove more than 20-30 µl, and make sure that the array of pores are covered by buffer at all times. Introducing air bubbles into the array can irreversibly damage pores.
After opening the inlet port, draw back a small volume to remove any air bubbles:
- Set a P1000 pipette tip to 200 µl.
- Insert the tip into the inlet port.
- Turn the wheel until the dial shows 220-230 µl, or until you see a small volume of buffer entering the pipette tip.

Slowly load 500 µl of the priming mix into the inlet port, as follows:
- Using a P1000 pipette, take 500 µl of the priming mix
- Insert the pipette tip into the priming port, ensuring there are no bubbles in the tip
- Slowly twist the pipette wheel down to load the flow cell (if possible with your pipette) or push down the plunger very slowly, leaving a small volume of buffer in the pipette tip.

It is vital that the inlet port is closed before removing waste to prevent air from being drawn across the sensor array area, which would lead to a significant loss of sequencing channels.
Remove the waste buffer, as follows:
- Ensure the inlet port is closed.
- Insert a P1000 pipette into a waste port and remove the waste buffer
Note: As the inlet port is closed, no fluid should leave the sensor array area.
Slide the inlet port cover clockwise to open.

Take care when drawing back buffer from the flow cell. Do not remove more than 20-30 µl, and make sure that the array of pores are covered by buffer at all times. Introducing air bubbles into the array can irreversibly damage pores.
After opening the inlet port, draw back a small volume to remove any air bubbles:
- Set a P1000 pipette tip to 200 µl.
- Insert the tip into the inlet port.
- Turn the wheel until the dial shows 220-230 µl, or until you see a small volume of buffer entering the pipette tip.

Mix the prepared library gently by pipetting up and down just prior to loading.
Load 200 µl of library into the inlet port using a P1000 pipette.

Close the valve to seal the inlet port.
Install the light shield on your flow cell as soon as library has been loaded for optimal sequencing output.
We recommend leaving the light shield on the flow cell when library is loaded, including during any washing and reloading steps. The shield can be removed when the library has been removed from the flow cell.
If the light shield has been removed from the flow cell, install the light shield as follows:
- Align the inlet port cut out of the light shield with the inlet port cover on the flow cell. The leading edge of the light shield should sit above the flow cell ID.
- Firmly press the light shield around the inlet port cover. The inlet port clip will click into place underneath the inlet port cover.


Close the PromethION lid when ready to start a sequencing run on MinKNOW.
Wait a minimum of 10 minutes after loading the flow cells onto the PromethION before initiating any experiments. This will help to increase the sequencing output.
Resume the sequencing run in MinKNOW to continue data acquisition.
After the final Pore-C DNA library has been loaded and sequenced, the flow cell can be flushed with deionised water and returned to Oxford Nanopore. For further information, see the "Ending the experiment" step.
26. Duplex experiment: Data acquisition and basecalling
How to start sequencing duplex data
The sequencing device control, data acquisition and real-time simplex basecalling are carried out by the MinKNOW software. After simplex data has been basecalled, duplex basecalling is performed on Dorado, which is detailed below the MinKNOW settings.
Ensure you are using the most recent software version of MinKNOW.
We recommend basecalling with the Fast basecaller in real-time using the PromethION 24 or 48 device. After simplex basecalling on MinKnow, the data can be rebasecalled using the super accurate (SUP) model when performing duplex basecalling on Dorado.
For detailed instructions for setting up the device and sequencing run, see the MinKNOW protocol section for "Starting a sequencing run with PromethION 24 and 48".
For further detailed instructions for simplex and duplex basecalling, see the Kit 14 sequencing and duplex basecalling document.
MinKNOW settings for the Duplex experiment
For the Duplex experiment, we recommend keeping the sequencing parameters to their default settings for simplex basecalling.
Below are the current recommendations:
Positions
Flow cell position: [User defined] Experiment name: [User defined] Flow cell type: FLO-PRO114M Sample ID: [User defined]

Kit selection:
Kit selection: Ligation Sequencing Kit XL V14 (SQK-LSK114-XL)

Run options:
Run duration: 100 hours Minimum read length: 200 bp
Adaptive sampling
Enrich or deplete sequencing: Off Barcoding balance: Not applicable
Advanced options
Active channel selection: On Time between pore scans: 1.5 hours Reserve pores: On

Analysis:
Basecalling
Basecalling: On Modified bases: On with '5mC' selected Model: Fast basecalling
Barcoding
Barcoding: Disabled
Alignment
Reference sequence: Off Note: We do not currently recommend live alignment during sequencing as it can slow down system processing.

Output:
Output format
Raw reads: On .FAST5: Off .POD5: On .FASTQ: On .BAM: On

Filtering: On Qscore: 8 Read length: Unfiltered Read splitting: Enabled Override read splitting min score: On

Overview of performing duplex basecalling:
Set up sequencing parameters in MinKNOW to perform simplex basecalling as described in "Basecalling Kit 14 simplex data". a. Basecall using the Fast basecaller. b. Output .POD5 files.
Using Dorado, re-basecall your simplex data with the following command to output simplex and duplex reads using the super-accurate (SUP) basecalling model. Other basecalling models can be used, as listed on the Dorado Github page.
$ dorado duplex dna_r10.4.1_e8.2_400bps_sup@v4.1.0 pod5s/ > duplex.bam
Dorado is a high-performance basecaller which is used to perform duplex basecalling. For further information about Dorado, please see the Dorado Github page.
Note: When running Dorado, we recommend stopping other basecalling for the best performance by maximising memory available to Dorado. This can be stopped and restarted when Dorado has finished via the GUI on MinKNOW.
Guppy may also be used to duplex basecall.
27. Ultra-long DNA experiment: Data acquisition and basecalling
How to start sequencing ultra-long read data
The sequencing device control, data acquisition and real-time simplex basecalling are carried out by the MinKNOW software.
Ensure you are using the most recent software version of MinKNOW.
We recommend basecalling with the high accuracy (HAC) basecaller in real-time using the PromethION 24 or 48 device. After basecalling, the simplex data can be rebasecalled using the super accurate (SUP) model and aligned to a reference genome.
For detailed instructions for setting up the device and sequencing run, see the MinKNOW protocol section for "Starting a sequencing run with PromethION 24 and 48".
MinKNOW settings for the Ultra-long DNA experiment
For the Ultra-long DNA experiment, we recommend keeping the sequencing parameters to their default settings for simplex basecalling.
Below are the current recommendations:
Poisitions
Flow cell position: [User defined]
Experiment name: [User defined]
Flow cell type: FLO-PRO114M
Sample ID: [User defined]

Kit selection:
Kit selection: Ultra-Long DNA Sequencing Kit V14 (SQK-LSK114)

Sequencing speed: Default (400 bps)

Run options:
Run duration: 72 hours
Minimum read length: 1000 bp
Adaptive sampling
Enrich or deplete sequencing: Off Barcoding balance: Not applicable
Advanced options
Active channel selection: On Time between pore scans: 1.5 hours Reserve pores: On

Analysis:
Basecalling
Basecalling: On Modified bases: On with '5mC' selected Model: High-accuracy basecalling (HAC)
Barcoding
Barcoding: Disabled
Alignment
Reference sequence: Off Note: We do not currently recommend live alignment during sequencing as it can slow down system processing.

Output:
Output format
Raw reads: On .FAST5: On .POD5: Off .FASTQ: On .BAM: On

Filtering: On Qscore: 9 Read length: Unfiltered Read splitting: Enabled Override read splitting min score: On

28. Pore-C experiment: Data acquisition and basecalling
How to start sequencing Pore-C data
The sequencing device control, data acquisition and real-time simplex basecalling are carried out by the MinKNOW software.
Ensure you are using the most recent software version of MinKNOW.
We recommend basecalling using the high accuracy (HAC) basecaller in real-time using the PromethION 24 or 48 device. After basecalling, the simplex data can be rebasecalled using the super accurate (SUP) model and aligned to a reference genome.
For detailed instructions for setting up the device and sequencing run, see the MinKNOW protocol section for "Starting a sequencing run with PromethION 24 and 48".
For further detailed instructions for simplex and duplex basecalling, see the Kit 14 sequencing and duplex basecalling document.
MinKNOW settings for the Pore-C experiment
For the Pore-C experiment, we recommend keeping the sequencing parameters to their default settings for simplex basecalling.
Below are the current recommendations:
Poisitions
Flow cell position: [User defined]
Experiment name: [User defined]
Flow cell type: FLO-PRO114M
Sample ID: [User defined]

Kit selection:
Kit selection: Ligation Sequencing Kit XL V14 (SQK-LSK114-XL)

Sequencing speed: Default (400 bps)
Run options:
Run duration: 72 hours
Minimum read length: 200 bp
Adaptive sampling
Enrich or deplete sequencing: Off Barcoding balance: Not applicable
Advanced options
Active channel selection: On Time between pore scans: 1.5 hours Reserve pores: On

Analysis:
Basecalling
Basecalling: On Modified bases: On with '5mC' selected Model: High-accuracy basecalling (HAC)
Barcoding
Barcoding: Disabled
Alignment
Reference sequence: Off Note: We do not currently recommend live alignment during sequencing as it can slow down system processing.

Output:
Output format
Raw reads: On .FAST5: On .POD5: Off .FASTQ: On .BAM: On

Filtering: On Qscore: 9 Read length: Unfiltered Read splitting: Enabled Override read splitting min score: On

29. Downstream analysis
Telomere-to-telomere assembly
We currently recommend users with bioinformatic experience to analyse their data using Verkko, a genome assembly pipeline specifically designed for large, eukaryotic genomes and incorporates duplex data (high accuracy reads) and ultra-long reads for assembly of a human chromosome. This workflow starts by building a multiplex de Bruijn graph based on the duplex data and uses the ultra-long reads to simplify the graph for assembly of a chromosome.
To provide functionality for the detection of Pore-C ligation junctions and the genomic assembly of fragments, we recommend using Pore-C-Snakemake and Pore-C tools which will be made available through our EPI2ME products.
We will be releasing an EPI2ME workflow combining the three datasets for more accessibilty for users of all experience. Further information will be released on the Community.
30. Flow cell reuse and returns
材料
- 测序芯片清洗剂盒(EXP-WSH004)
After your sequencing experiment is complete, if you would like to reuse the flow cell, please follow the Flow Cell Wash Kit protocol and store the washed flow cell at +2°C to +8°C.
The Flow Cell Wash Kit protocol is available on the Nanopore Community.
We recommend you to wash the flow cell as soon as possible after you stop the run. However, if this is not possible, leave the flow cell on the device and wash it the next day.
Alternatively, follow the returns procedure to send the flow cell back to Oxford Nanopore.
Instructions for returning flow cells can be found here.
If you encounter issues or have questions about your sequencing experiment, please refer to the Troubleshooting Guide that can be found in the online version of this protocol.
31. Issues during DNA/RNA extraction and library preparation
Below is a list of the most commonly encountered issues, with some suggested causes and solutions.
We also have an FAQ section available on the Nanopore Community Support section.
If you have tried our suggested solutions and the issue still persists, please contact Technical Support via email (support@nanoporetech.com) or via LiveChat in the Nanopore Community.
Low sample quality
| Observation | Possible cause | Comments and actions |
|---|---|---|
| Low DNA purity (Nanodrop reading for DNA OD 260/280 is <1.8 and OD 260/230 is <2.0–2.2) | The DNA extraction method does not provide the required purity | The effects of contaminants are shown in the Contaminants document. Please try an alternative extraction method that does not result in contaminant carryover. Consider performing an additional SPRI clean-up step. |
| Low RNA integrity (RNA integrity number <9.5 RIN, or the rRNA band is shown as a smear on the gel) | The RNA degraded during extraction | Try a different RNA extraction method. For more info on RIN, please see the RNA Integrity Number document. Further information can be found in the DNA/RNA Handling page. |
| RNA has a shorter than expected fragment length | The RNA degraded during extraction | Try a different RNA extraction method. For more info on RIN, please see the RNA Integrity Number document. Further information can be found in the DNA/RNA Handling page. We recommend working in an RNase-free environment, and to keep your lab equipment RNase-free when working with RNA. |
Low DNA recovery after AMPure bead clean-up
| Observation | Possible cause | Comments and actions |
|---|---|---|
| Low recovery | DNA loss due to a lower than intended AMPure beads-to-sample ratio | 1. AMPure beads settle quickly, so ensure they are well resuspended before adding them to the sample. 2. When the AMPure beads-to-sample ratio is lower than 0.4:1, DNA fragments of any size will be lost during the clean-up. |
| Low recovery | DNA fragments are shorter than expected | The lower the AMPure beads-to-sample ratio, the more stringent the selection against short fragments. Please always determine the input DNA length on an agarose gel (or other gel electrophoresis methods) and then calculate the appropriate amount of AMPure beads to use. 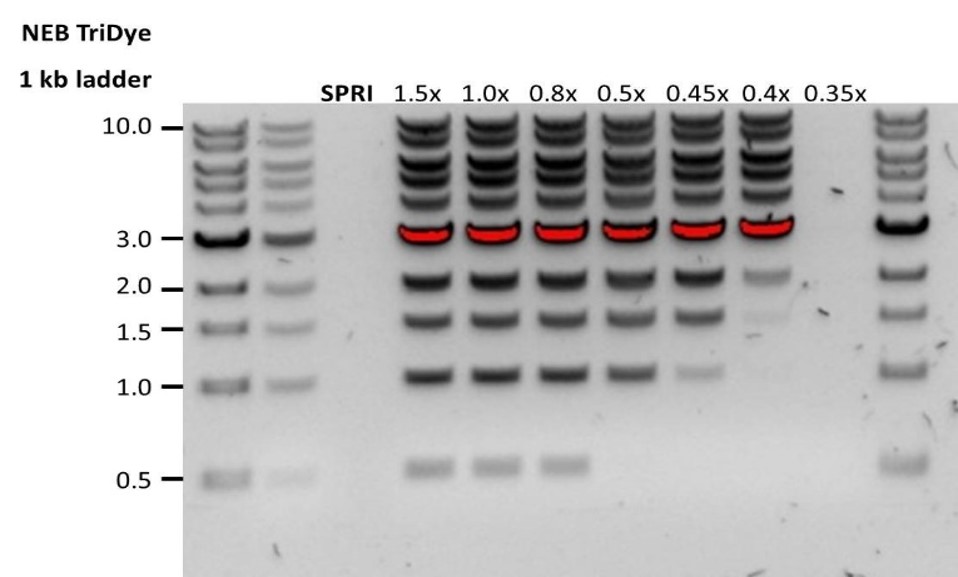 |
| Low recovery after end-prep | The wash step used ethanol <70% | DNA will be eluted from the beads when using ethanol <70%. Make sure to use the correct percentage. |
32. Issues during the sequencing run
Below is a list of the most commonly encountered issues, with some suggested causes and solutions.
We also have an FAQ section available on the Nanopore Community Support section.
If you have tried our suggested solutions and the issue still persists, please contact Technical Support via email (support@nanoporetech.com) or via LiveChat in the Nanopore Community.
Fewer pores at the start of sequencing than after Flow Cell Check
| Observation | Possible cause | Comments and actions |
|---|---|---|
| MinKNOW reported a lower number of pores at the start of sequencing than the number reported by the Flow Cell Check | An air bubble was introduced into the nanopore array | After the Flow Cell Check it is essential to remove any air bubbles near the priming port before priming the flow cell. If not removed, the air bubble can travel to the nanopore array and irreversibly damage the nanopores that have been exposed to air. The best practice to prevent this from happening is demonstrated in this video. |
| MinKNOW reported a lower number of pores at the start of sequencing than the number reported by the Flow Cell Check | The flow cell is not correctly inserted into the device | Stop the sequencing run, remove the flow cell from the sequencing device and insert it again, checking that the flow cell is firmly seated in the device and that it has reached the target temperature. If applicable, try a different position on the device (GridION/PromethION). |
| MinKNOW reported a lower number of pores at the start of sequencing than the number reported by the Flow Cell Check | Contaminations in the library damaged or blocked the pores | The pore count during the Flow Cell Check is performed using the QC DNA molecules present in the flow cell storage buffer. At the start of sequencing, the library itself is used to estimate the number of active pores. Because of this, variability of about 10% in the number of pores is expected. A significantly lower pore count reported at the start of sequencing can be due to contaminants in the library that have damaged the membranes or blocked the pores. Alternative DNA/RNA extraction or purification methods may be needed to improve the purity of the input material. The effects of contaminants are shown in the Contaminants Know-how piece. Please try an alternative extraction method that does not result in contaminant carryover. |
MinKNOW script failed
| Observation | Possible cause | Comments and actions |
|---|---|---|
| MinKNOW shows "Script failed" | Restart the computer and then restart MinKNOW. If the issue persists, please collect the MinKNOW log files and contact Technical Support. If you do not have another sequencing device available, we recommend storing the flow cell and the loaded library at 4°C and contact Technical Support for further storage guidance. |
Pore occupancy below 40%
| Observation | Possible cause | Comments and actions |
|---|---|---|
| Pore occupancy <40% | Not enough library was loaded on the flow cell | Ensure the correct volume and concentration as stated on the appropriate protocol for your sequencing library is loaded onto the flow cell. Please quantify the library before loading and calculate fmols using tools like the Promega Biomath Calculator, choosing "dsDNA: µg to fmol" |
| Pore occupancy close to 0 | The Ligation Sequencing Kit was used, and sequencing adapters did not ligate to the DNA | Make sure to use the NEBNext Quick Ligation Module (E6056) and Oxford Nanopore Technologies Ligation Buffer (LNB, provided in the sequencing kit) at the sequencing adapter ligation step, and use the correct amount of each reagent. A Lambda control library can be prepared to test the integrity of the third-party reagents. |
| Pore occupancy close to 0 | The Ligation Sequencing Kit was used, and ethanol was used instead of LFB or SFB at the wash step after sequencing adapter ligation | Ethanol can denature the motor protein on the sequencing adapters. Make sure the LFB or SFB buffer was used after ligation of sequencing adapters. |
| Pore occupancy close to 0 | No tether on the flow cell | Tethers are adding during flow cell priming (FLT tube for Kit 9, 10, 11, FCT for Kit 14, and FTU for ultra-long DNA kits). Make sure FLT/FCT/FTU was added to the buffer (FB for Kit 9, 10, 11, and FCF for Kit 14) before priming. |
Shorter than expected read length
| Observation | Possible cause | Comments and actions |
|---|---|---|
| Shorter than expected read length | Unwanted fragmentation of DNA sample | Read length reflects input DNA fragment length. Input DNA can be fragmented during extraction and library prep. 1. Please review the Extraction Methods in the Nanopore Community for best practice for extraction. 2. Visualise the input DNA fragment length distribution on an agarose gel before proceeding to the library prep.  In the image above, Sample 1 is of high molecular weight, whereas Sample 2 has been fragmented. In the image above, Sample 1 is of high molecular weight, whereas Sample 2 has been fragmented.3. During library prep, avoid pipetting and vortexing when mixing reagents. Flicking or inverting the tube is sufficient. |
Large proportion of unavailable pores
| Observation | Possible cause | Comments and actions |
|---|---|---|
Large proportion of unavailable pores (shown as blue in the channels panel and pore activity plot) 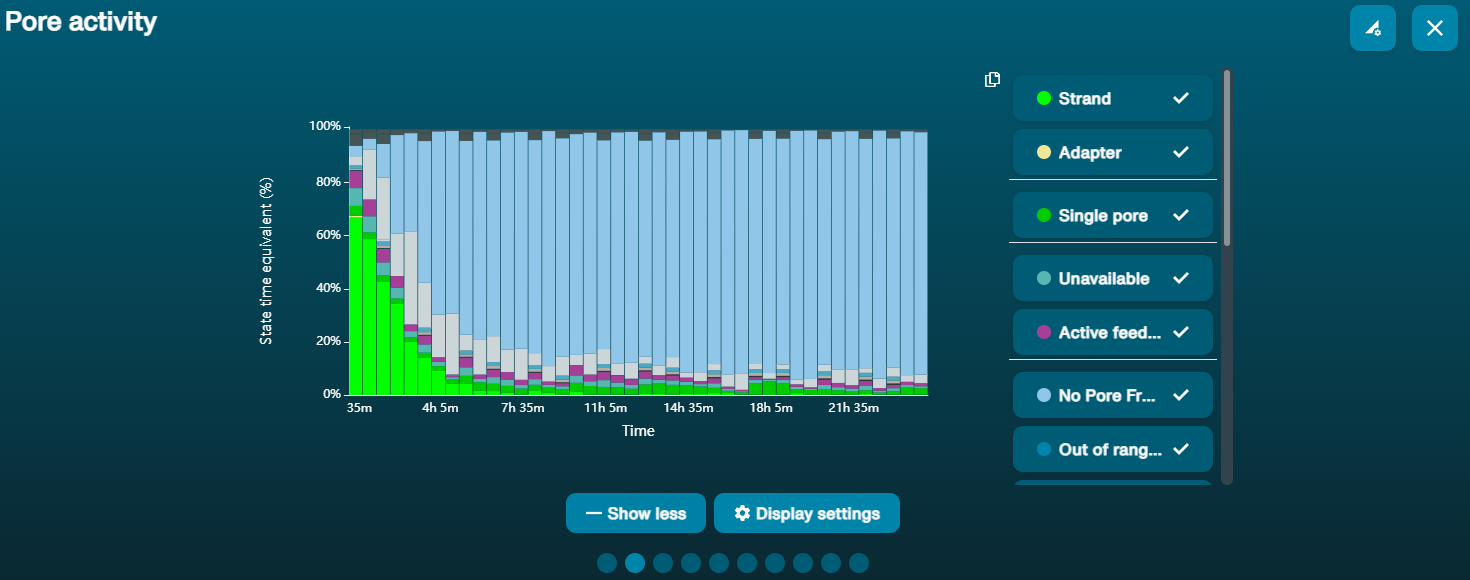 The pore activity plot above shows an increasing proportion of "unavailable" pores over time. The pore activity plot above shows an increasing proportion of "unavailable" pores over time. | Contaminants are present in the sample | Some contaminants can be cleared from the pores by the unblocking function built into MinKNOW. If this is successful, the pore status will change to "sequencing pore". If the portion of unavailable pores stays large or increases: 1. A nuclease flush using the Flow Cell Wash Kit (EXP-WSH004) can be performed, or 2. Run several cycles of PCR to try and dilute any contaminants that may be causing problems. |
Large proportion of inactive pores
| Observation | Possible cause | Comments and actions |
|---|---|---|
| Large proportion of inactive/unavailable pores (shown as light blue in the channels panel and pore activity plot. Pores or membranes are irreversibly damaged) | Air bubbles have been introduced into the flow cell | Air bubbles introduced through flow cell priming and library loading can irreversibly damage the pores. Watch the Priming and loading your flow cell video for best practice |
| Large proportion of inactive/unavailable pores | Certain compounds co-purified with DNA | Known compounds, include polysaccharides, typically associate with plant genomic DNA. 1. Please refer to the Plant leaf DNA extraction method. 2. Clean-up using the QIAGEN PowerClean Pro kit. 3. Perform a whole genome amplification with the original gDNA sample using the QIAGEN REPLI-g kit. |
| Large proportion of inactive/unavailable pores | Contaminants are present in the sample | The effects of contaminants are shown in the Contaminants Know-how piece. Please try an alternative extraction method that does not result in contaminant carryover. |
Temperature fluctuation
| Observation | Possible cause | Comments and actions |
|---|---|---|
| Temperature fluctuation | The flow cell has lost contact with the device | Check that there is a heat pad covering the metal plate on the back of the flow cell. Re-insert the flow cell and press it down to make sure the connector pins are firmly in contact with the device. If the problem persists, please contact Technical Services. |
Failed to reach target temperature
| Observation | Possible cause | Comments and actions |
|---|---|---|
| MinKNOW shows "Failed to reach target temperature" | The instrument was placed in a location that is colder than normal room temperature, or a location with poor ventilation (which leads to the flow cells overheating) | MinKNOW has a default timeframe for the flow cell to reach the target temperature. Once the timeframe is exceeded, an error message will appear and the sequencing experiment will continue. However, sequencing at an incorrect temperature may lead to a decrease in throughput and lower q-scores. Please adjust the location of the sequencing device to ensure that it is placed at room temperature with good ventilation, then re-start the process in MinKNOW. Please refer to this link for more information on MinION temperature control. |







