Flow Cell Wash Kit (EXP-WSH004 or EXP-WSH004-XL) (WFC_9120_v1_revS_25Jul2025)
PromethION: Protocol
V WFC_9120_v1_revS_25Jul2025
FOR RESEARCH USE ONLY
Contents
Introduction to the protocol
Prepare for your flow cell wash
- 3. Method considerations and identifying your PromethION Flow Cell components
- 4. Prepare your Flow Cell Wash Kit reagents
Wash your flow cell
Post-wash options
1. Overview of the protocol
Introduction to the Flow Cell Wash Kit
The Flow Cell Wash Kit allows sequential runs of sequencing libraries on the same flow cell. It works by flushing out and digesting the DNA library currently on the flow cell, and refreshing the system for a subsequent or new library to be loaded. The Flow Cell Wash Kit is compatible with R9.4.1 and R10.4.1 flow cells.
The Flow Cell Wash Kit can also be used with our RNA flow cells to flush out RNA libraries, but will not remove RNA-related blocking and restore nanopores.
Using the Flow Cell Wash Kit provides the opportunity to utilise the same flow cell a number of times*, maximising the available run time on your flow cell.
*The number of Flow Cell washes and reuse will depend on various factors. These numbers typically range from 3-6 flow cell wash and reuses. For more information on flow cell reuse and limiting factors please refer to our Flow Cell Wash Kit (EXP-WSH004 or EXP-WSH004-XL) know-how document.
Following the wash step you have two options for reusing your flow cell:
- a subsequent or new library can be loaded on your washed flow cell to begin sequencing straight away.
- Storage Buffer (S) from the Flow Cell Wash Kit can be introduced into the flow cell, allowing you to store the washed flow cell at 2–8°C for later use.
Flow Cell Wash Kit (EXP-WSH004 or EXP-WSH004-XL) know-how document
For more information on how the Flow Cell Wash Kit (EXP-WSH004 or EXP-WSH004-XL) works, performance data and additional recommendations; please see our Flow Cell Wash Kit (EXP-WSH004 or EXP-WSH004-XL) know-how document.
2. Equipment and consumables
材料
- 测序芯片清洗剂盒(EXP-WSH004)或测序芯片清洗试剂盒 XL(EXP-WSH004-XL)
- Flow cell priming reagents available in your sequencing kit or in the following kits:
- 测序辅助扩展包 V14(EXP-AUX003)
- Flow Cell Priming Kit (EXP-FLP004)
仪器
- P1000 移液枪和枪头
- P20 移液枪和枪头
- 盛有冰的冰桶
Reloading a library
Additional buffers may be required for reloading a library following the washing of a flow cell. These can be found in the one of the following expansion kits:
Sequencing Auxiliary Vials V14 (EXP-AUX003). This expansion contains vials of Sequencing Buffer (SB), Elution Buffer (EB), Library Solution (LIS), and Library Beads (LIB) with Kit 14 flow cell priming reagents: Flow Cell Flush (FCF) and Flow Cell Tether (FCT). This expansion is only compatible with our Kit 14 chemistry.
Flow Cell Priming Kit (EXP-FLP004). This expansion contains both Kit 14 flow cell priming reagents required: Flow Cell Flush (FCF) and Flow Cell Tether (FCT). This expansion is only compatible with Kit 14 chemistry.
Flow Cell Wash Kit (EXP-WSH004) contents
- Wash Mix (WMX) contains DNase I.
- Wash Diluent (DIL) contains the exonuclease buffer that maximises activity of the DNase I.
- The Storage Buffer allows flow cells to be stored until their next use.
Flow Cell Wash Kit XL (EXP-WSH004-XL) contents
- Wash Mix (WMX) contains DNase I.
- Wash Diluent (DIL) contains the exonuclease buffer that maximises activity of the DNase I.
- The Storage Buffer allows flow cells to be stored until their next use.
3. Method considerations and identifying your PromethION Flow Cell components
Keep your flow cell on your sequencing device during the whole wash procedure.
We strongly recommend your flow cell is kept inserted in the sequencing device to ensure the equipment is secured and to maintain proper temperature control throughout the wash procedure.
Flow cell light shield during flow cell wash.
The flow cell light shield can be kept on the flow cell during the flow cell wash method. However, if you would prefer to remove it for visibility of the array this can be done.
Please ensure you follow protocol guidance and the flow cell light shield is replaced on the flow cell before resuming or starting a new sequencing run.
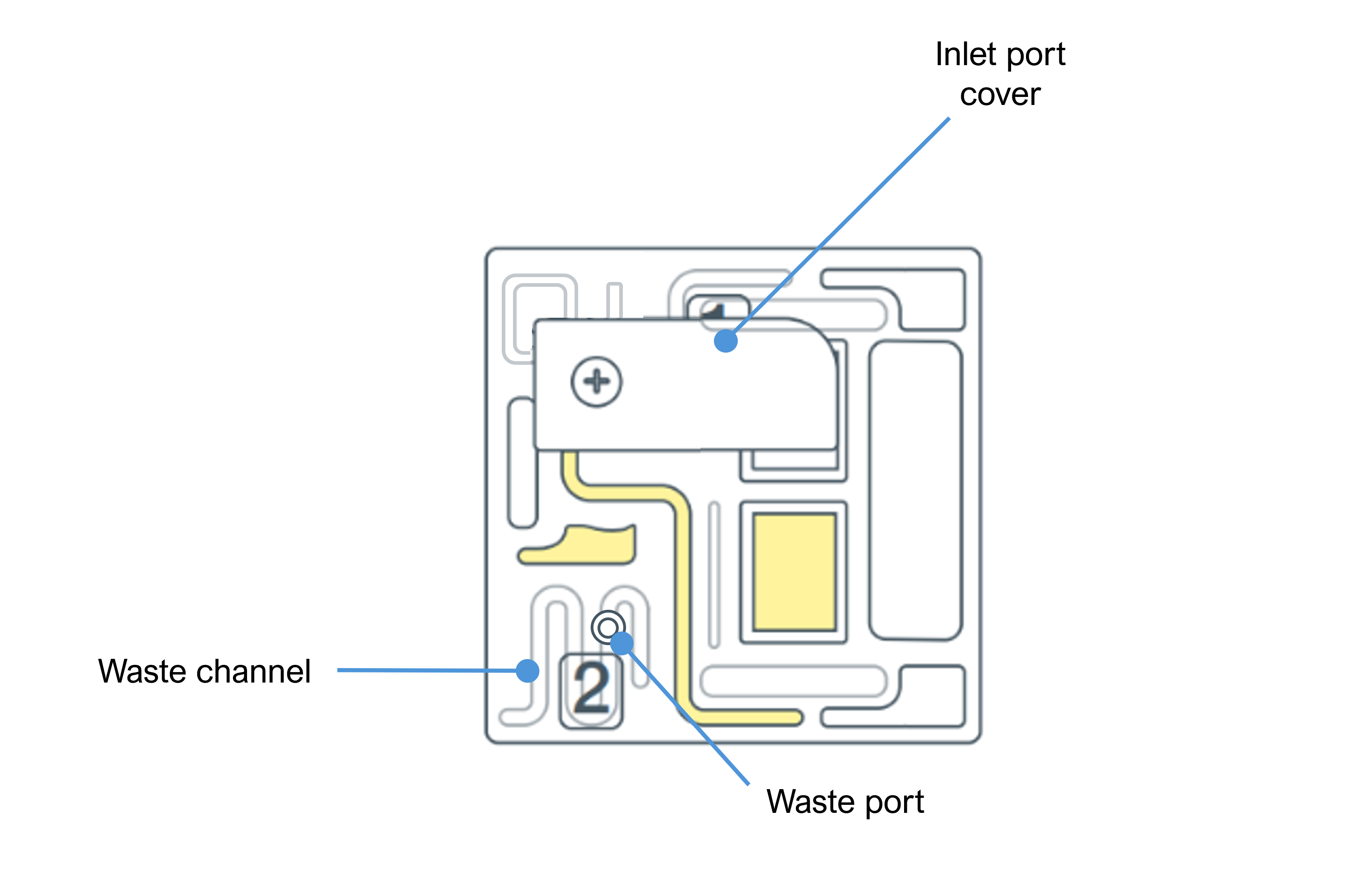
Familiarise yourself with the PromethION Flow Cell components.
If required, perform library recovery from your flow cell(s).
If you are planning on recovering your current sequencing library from your flow cell before proceeding with the flow cell wash, please ensure this is the first thing you do.
Guidance on how to recover your library from your flow cell can be found in our library recovery from flow cells protocol.
Watch the washing and reloading a PromethION Flow Cell video.
This video will show you how to wash a flow cell after a sequencing run and how to load a new library. We strongly recommend all our users to view the demonstration video before performing this method:
Washing and reloading a PromethION Flow Cell video
4. Prepare your Flow Cell Wash Kit reagents
材料
- 测序芯片清洗剂盒(EXP-WSH004)或测序芯片清洗试剂盒 XL(EXP-WSH004-XL)
耗材
- 1.5 ml Eppendorf DNA LoBind 离心管
仪器
- P1000 移液枪和枪头
- P20 移液枪和枪头
- 涡旋混匀仪
- 迷你离心机
- 盛有冰的冰桶
We strongly recommend preparing your flow cell wash reagents fresh for each use.
Do not pre-prepare your Flow Cell Wash Mix and store it for >1 day, as this could result in sub-optimal results.
Place the tube of Wash Mix (WMX) on ice. Do not vortex the tube.
Thaw one tube of Wash Diluent (DIL) at room temperature.
Mix the contents of Wash Diluent (DIL) thoroughly by vortexing, then spin down briefly and place on ice.
In a fresh 1.5 ml Eppendorf DNA LoBind tube, prepare the following Flow Cell Wash Mix:
| Reagent | Volume required per flow cell |
|---|---|
| Wash Mix (WMX) | 2 μl |
| Wash Diluent (DIL) | 398 μl |
| Total | 400 μl |
Mix well by pipetting, and place the tube(s) on ice.
Note: Do not vortex the Flow Cell Wash Mix tube.
Store the prepared Flow Cell Wash Mix tube on ice until required.
5. Pause/stop the sequencing run and remove the waste buffer
仪器
- P1000 移液枪和枪头
Keep your flow cell on your sequencing device during the whole wash procedure.
We strongly recommend your flow cell is kept inserted in the sequencing device to ensure the equipment is secured, minimise component damage and potential errors, and maximise usability.
Stop or pause the sequencing experiment in MinKNOW
Please ensure your flow cell is not sequencing while performing any of the wash steps. Manipulating the flow cell while it is sequencing can results in damage and pore loss.
Ensure the flow cell inlet port cover is closed.
Note: It is vital that the flow cell inlet port is closed before removing the waste buffer to prevent air from being drawn across the sensor array area, which would lead to a significant loss of sequencing channels.
Remove all of the waste buffers from the waste port by using a P1000 pipette.
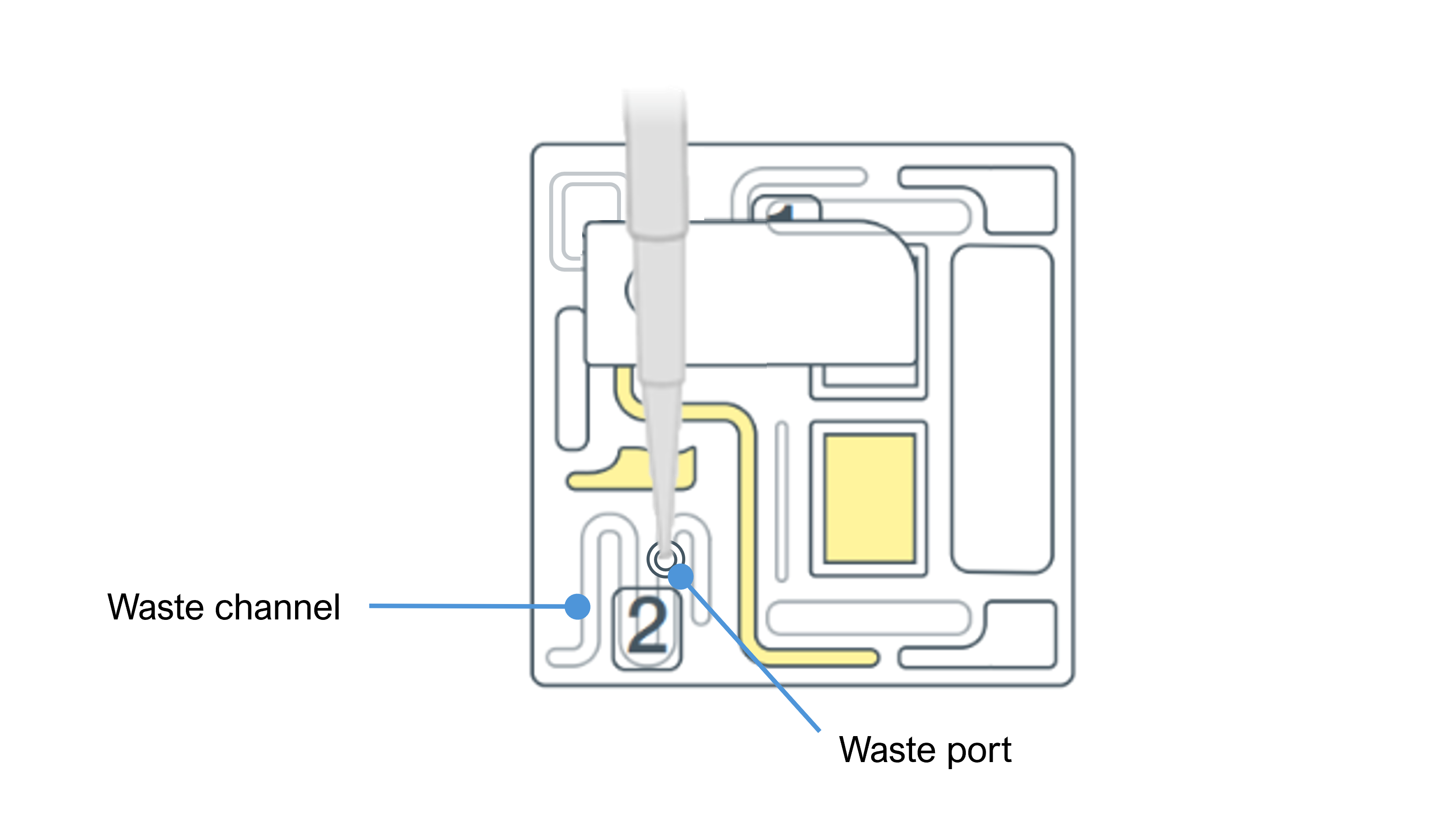
You should remove the full volume of waste buffer from the waste channel. If required you can dispense the volume aspirated from the waste channel and repeat the process to remove the full volume from the waste channel.
- Set your P1000 pipette to 1000 µl.
- Insert the P1000 pipette tip into waste port.
- Slowly aspirate to remove the waste buffer from waste port.
Note: As the inlet port is closed, no fluid should leave the sensor array area.
Proceed to wash your flow cell.
6. Wash the flow cell
材料
- Prepared Flow Cell Wash Mix (from previous step)
仪器
- P1000 移液枪和枪头
- P200 移液枪和枪头
- 盛有冰的冰桶
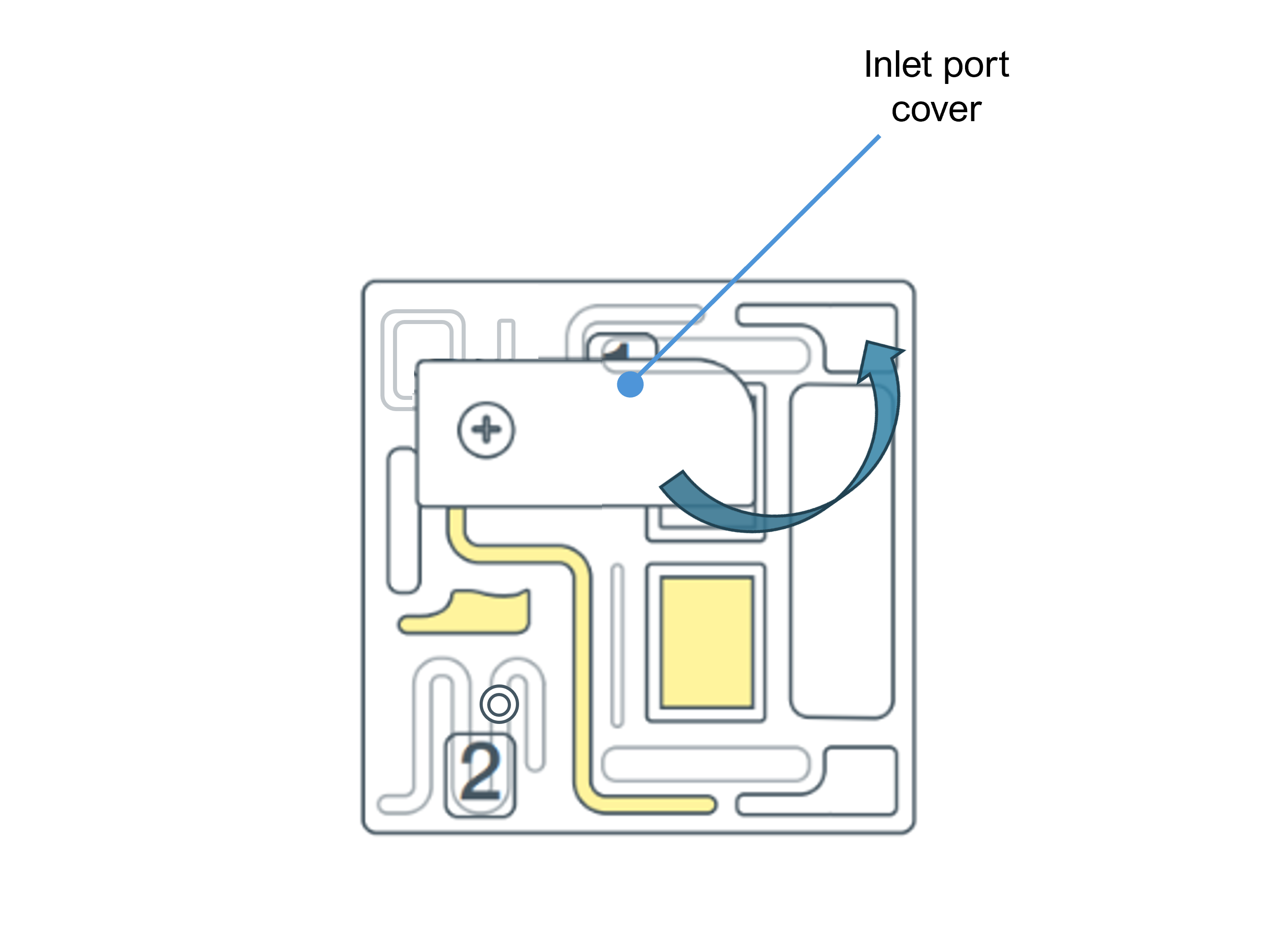
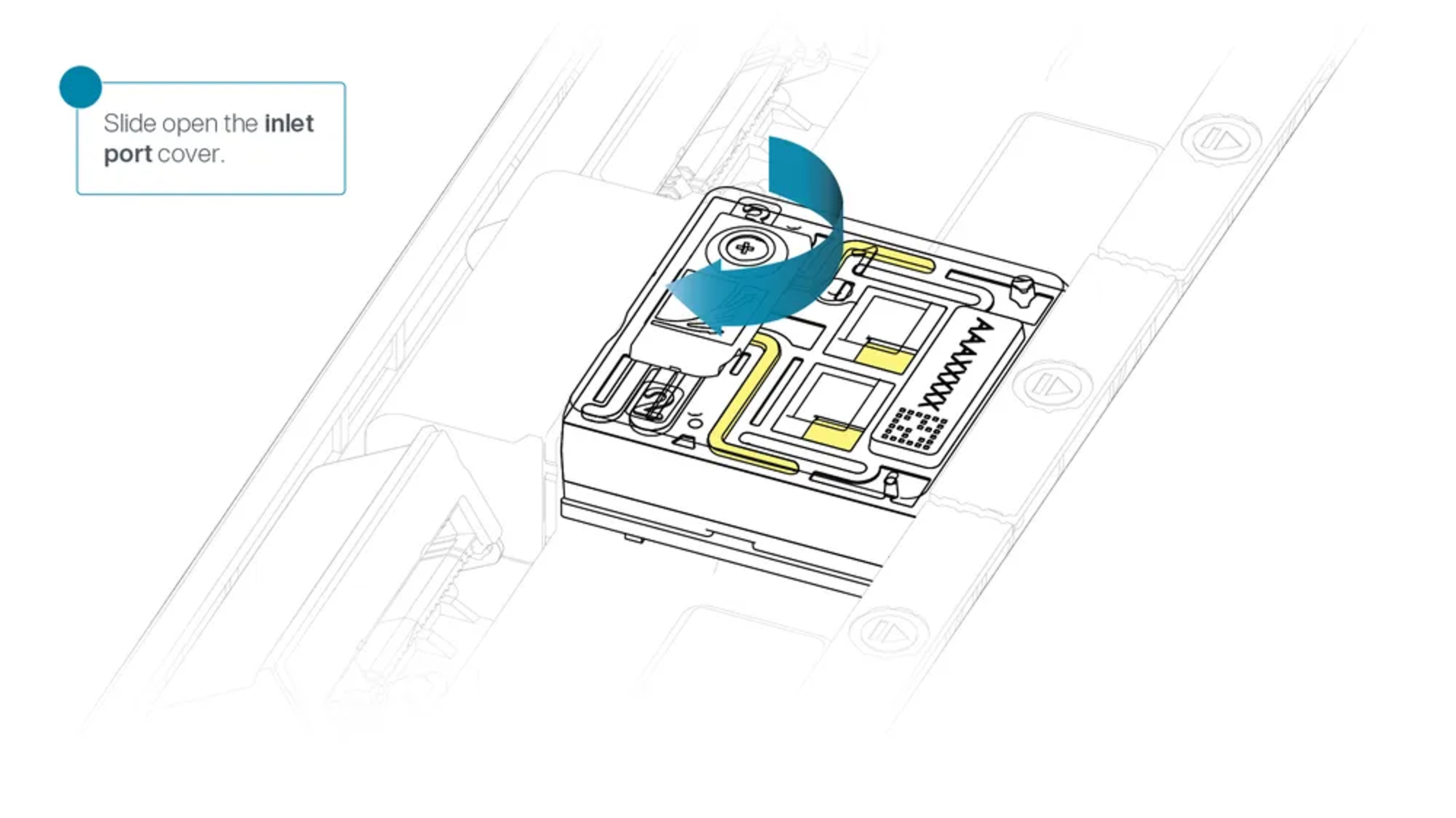
Slide the flow cell inlet port cover clockwise to open.
Take care when drawing back buffer from the flow cell. Do not remove more than 20-30 µl, and make sure that the array of pores are covered by buffer at all times. Introducing air bubbles into the array can irreversibly damage pores.
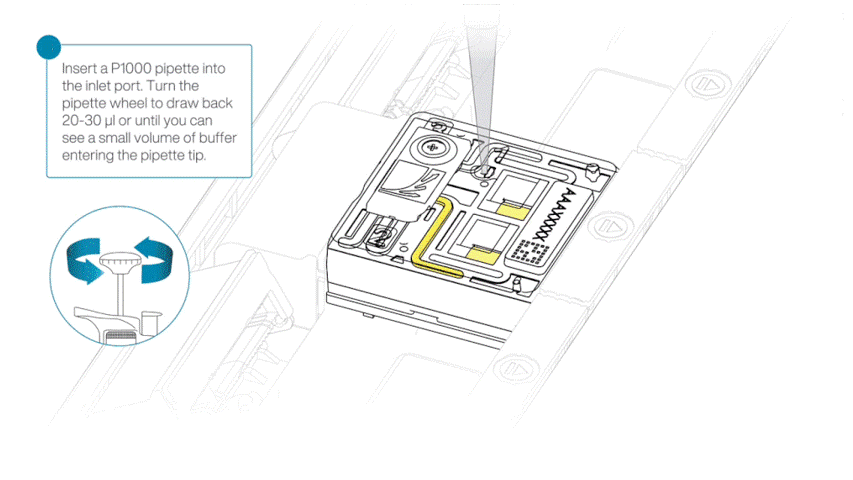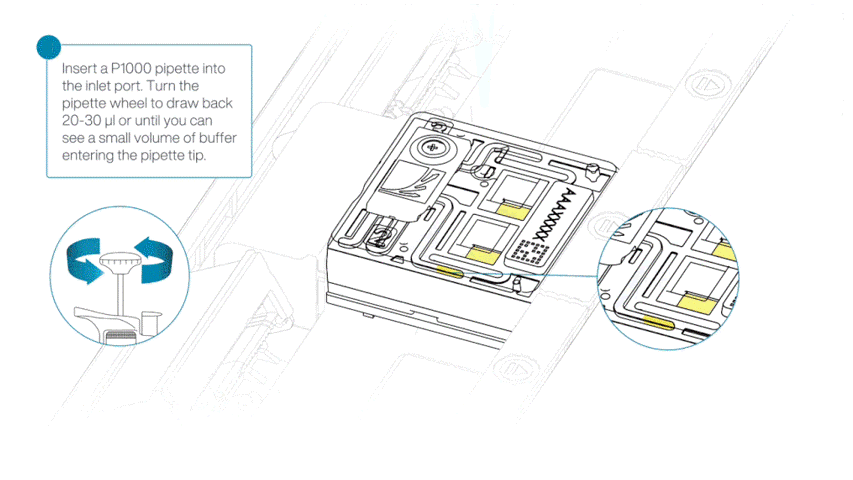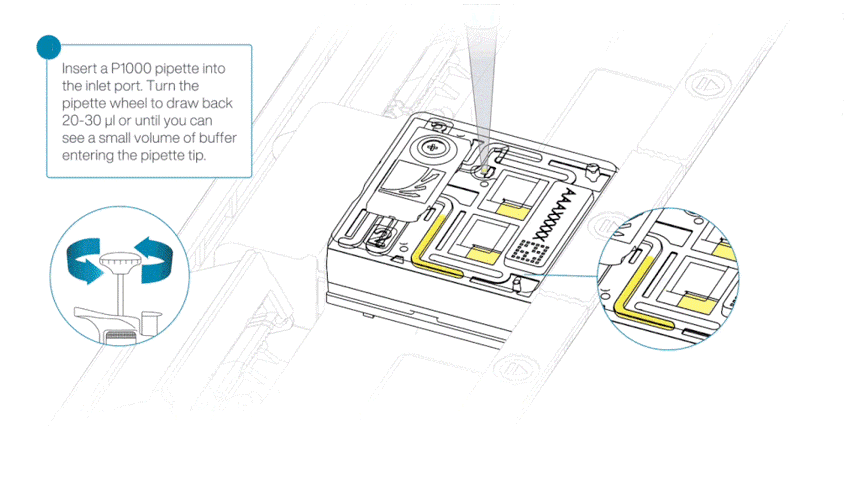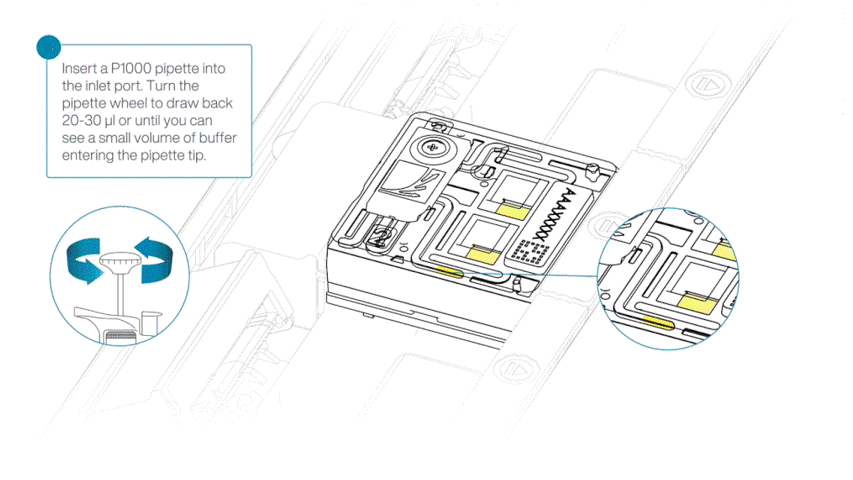
After opening the priming port, check for a small air bubble under the cover. Draw back a small volume to remove any bubbles:
- Set a P1000 pipette to 200 µl.
- Insert the tip into the flow cell priming port.
- Turn the wheel until the dial shows 220-230 µl, or until you can see a small volume of buffer/liquid entering the pipette tip.
- Visually check that there is continuous buffer from the flow cell priming port across the sensor array.
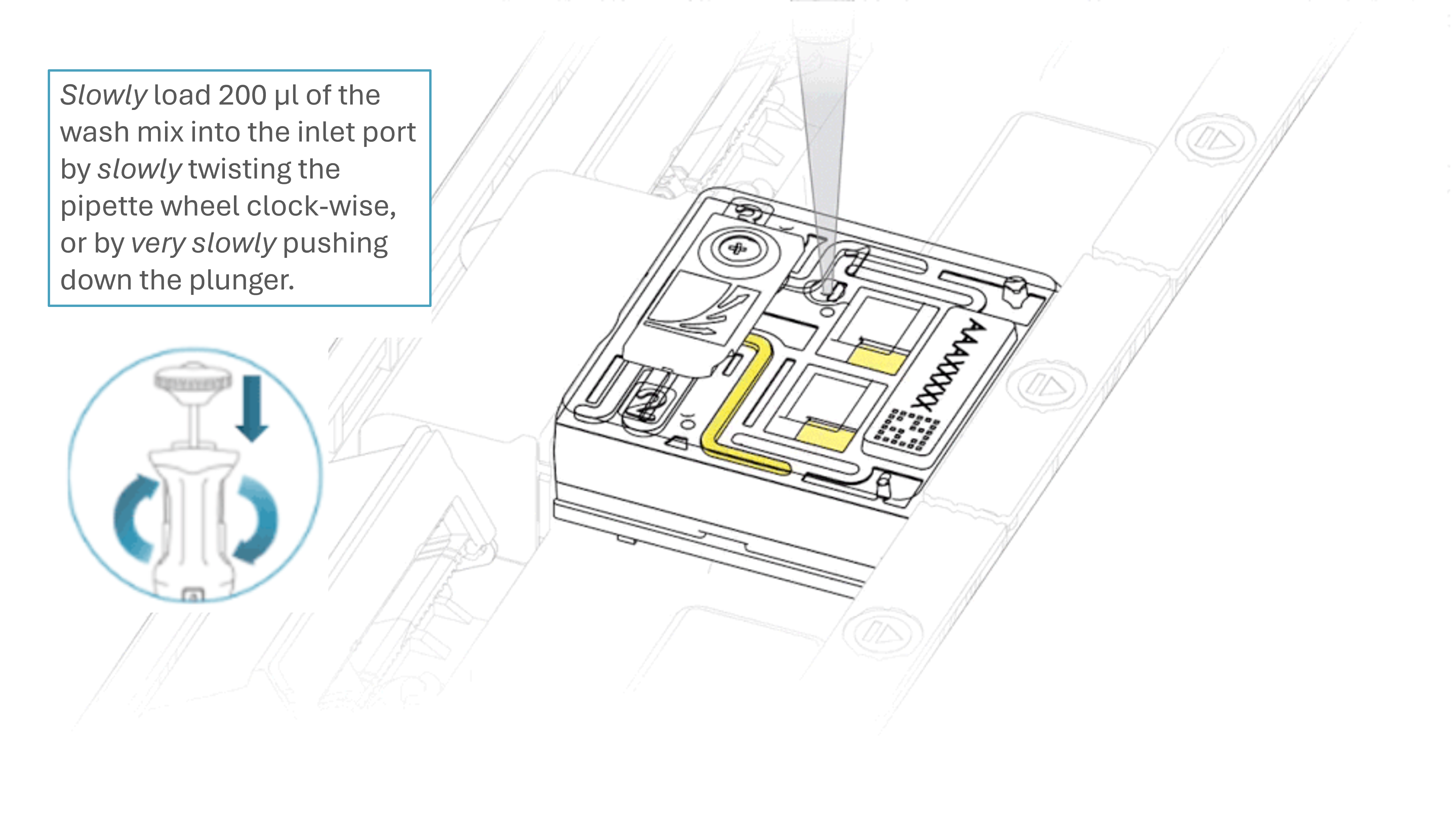
Slowly load 200 µl of the prepared flow cell wash mix into the inlet port, as follows:
- Using a P1000 pipette, take 200 µl of the flow cell wash mix
- Insert the pipette tip into the inlet port, ensuring there are no bubbles in the tip
- Slowly twist the pipette wheel down to load the flow cell (if possible with your pipette) or push down the plunger very slowly, leaving a small volume of buffer in the pipette tip.
Note: Dispensing the wash mix into your flow cell should take at least 5 seconds, ensuring it is done very slowly and at a continuous rate.
Set a timer and incubate the wash mix on your flow cell for 5 minutes.
Once the 5 minute incubation is complete, repeat the flow cell wash mix loading step (step 3), for a second 200 µl load of the prepared wash mix.
Close the inlet port and wait for 1 hour.

Remove all of the waste buffers from the waste port by using a P1000 pipette.

Your flow cell has now been washed and you can proceed to the next step.
Proceed to:
- Option 1: Run a second sequencing library on your flow cell
- Option 2: Store your flow cell for later use
Note: Do not store your flow cell with the wash mix on the array. This can lead to irreversible pore loss.
7. Option 1: Run a second sequencing library on your flow cell
材料
- Flow cell priming reagents available in your sequencing kit or in the following kits:
- 测序辅助扩展包 V14(EXP-AUX003)
- Flow Cell Priming Kit V14 (EXP-FLP004)
仪器
- P1000 移液枪和枪头
- P20 移液枪和枪头
- 盛有冰的冰桶
The sequencing reagents outlined in this method are for our most recent V14 chemisty.
If using a previous version of our chemistry or a kit with specific sequencing reagents, please ensure you are using the correct sequencing reagents for your application.
The buffers used in this process are incompatible with conducting a Flow Cell Check prior to loading a subsequent library. However, the first pore scan once a sequencing run has started will report the number of nanopores available.
Thaw the Sequencing Buffer (SB), Library Beads (LIB) or Library Solution (LIS, if using), Flow Cell Tether (FCT) and Flow Cell Flush (FCF) at room temperature before mixing by vortexing. Then spin down and store on ice.
Prepare the flow cell priming mix by combining the following reagents and thoroughly mixing by pipetteing:
| Reagent | Volume per flow cell |
|---|---|
| Flow Cell Flush (FCF) | 1,170 µl |
| Flow Cell Tether (FCT) | 30 µl |
| Total volume | 1,205 µl |
Slide the inlet port cover clockwise to open.
After opening the inlet port, check for a small air bubble under the cover. Draw back a small volume to remove any bubbles:
- Set a P1000 pipette to 200 µl
- Insert the tip into the inlet port
- Turn the wheel until the dial shows 220-230 µl, to draw back 20-30 µl, or until you can see a small volume of buffer entering the pipette tip
Note: Take care when drawing back buffer from the flow cell, do not remove more than 20-30 µl. Visually check that there is continuous buffer from the inlet port across the sensor array.
Slowly load 500 µl of the priming mix into the inlet port, as follows:
- Using a P1000 pipette, take 500 µl of the priming mix
- Insert the pipette tip into the inlet port, ensuring there are no bubbles in the tip
- Slowly twist the pipette wheel down to load the flow cell (if possible with your pipette) or push down the plunger very slowly, as illustrated in the demonstration video, leaving a small volume of buffer in the pipette tip.
Note: Dispensing the 500 µl of priming mix into your flow cell should take at least 5 seconds, ensuring it is done very slowly and at a continuous rate.
It is vital to wait five minutes between the priming mix flushes to effectively remove the nuclease.
Close the inlet port and wait five minutes.
During this time, prepare the library for loading by following the steps below.
Thoroughly mix the contents of the Library Beads (LIB) by pipetting.
The Library Beads (LIB) tube contains a suspension of beads. These beads settle very quickly. It is vital that they are mixed immediately before use.
We recommend using the Library Beads (LIB) for most sequencing experiments. However, the Library Solution (LIS) is available for more viscous libraries.
In a new tube, prepare the library for loading according to the "Priming and loading the PromethION Flow Cell" section of the suitable protocol to ensure you are using the correct reagents and volumes.
| Reagent | Volume per flow cell |
|---|---|
| Sequencing Buffer (SB) | 100 µl |
| Library Beads (LIB) thoroughly mixed before use, or Library Solution (LIS) | 68 µl |
| DNA library | 32 µl |
| Total | 200 µl |
Remove all of the waste buffers from the waste port by using a P1000 pipette.
You should remove the full volume of waste buffer from the waste channel. If required you can dispense the volume aspirated from the waste channel and repeat the process to remove the full volume from the waste channel.
- Set your P1000 pipette to 1000 µl.
- Insert the P1000 pipette tip into the waste port.
- Slowly aspirate to remove the waste buffer from the waste port.
Note: As the inlet port is closed, no fluid should leave the sensor array area.
Slowly, perform the second load of 500 µl of the priming mix into the inlet port, as follows:
- Open the inlet port.
- Using a P1000 pipette, take 500 µl of the priming mix
- Insert the pipette tip into the inlet port, ensuring there are no bubbles in the tip
- Slowly twist the pipette wheel down to load the flow cell (if possible with your pipette) or push down the plunger very slowly, as illustrated in the demonstration video, leaving a small volume of buffer in the pipette tip.
Note: Dispensing the 500 µl of priming mix into your flow cell should take at least 5 seconds, ensuring it is done very slowly and at a continuous rate.
Mix the prepared sequencing library gently by pipetting up and down just prior to loading.
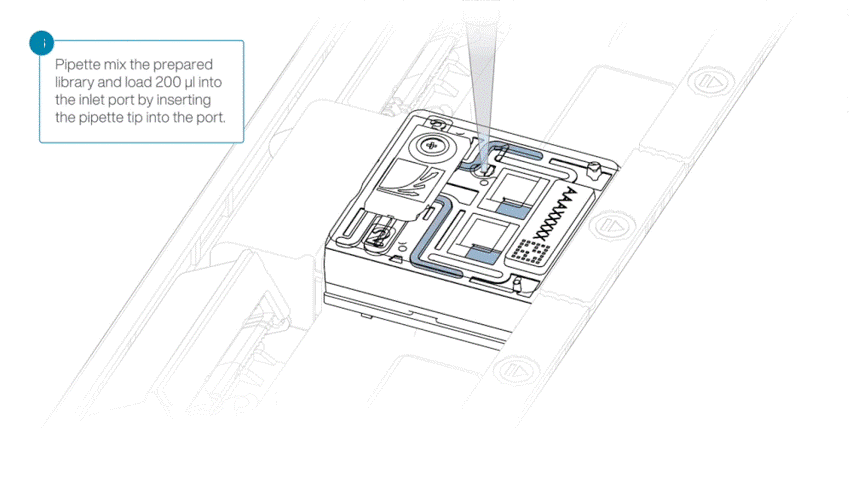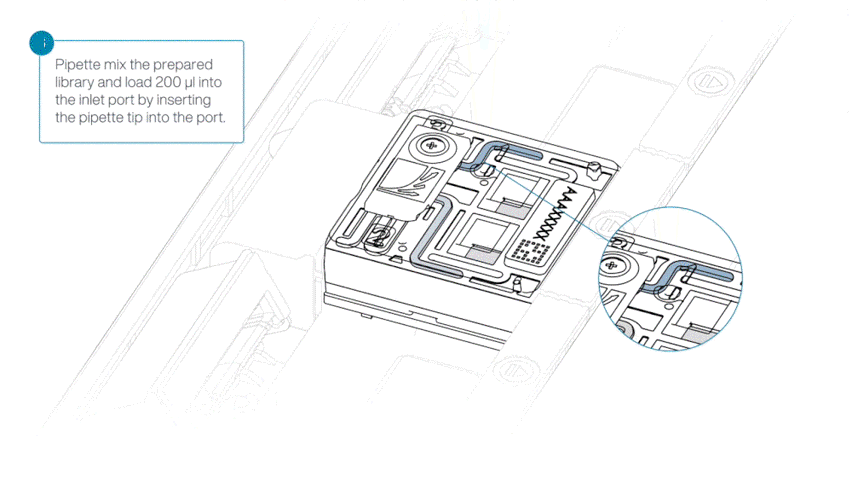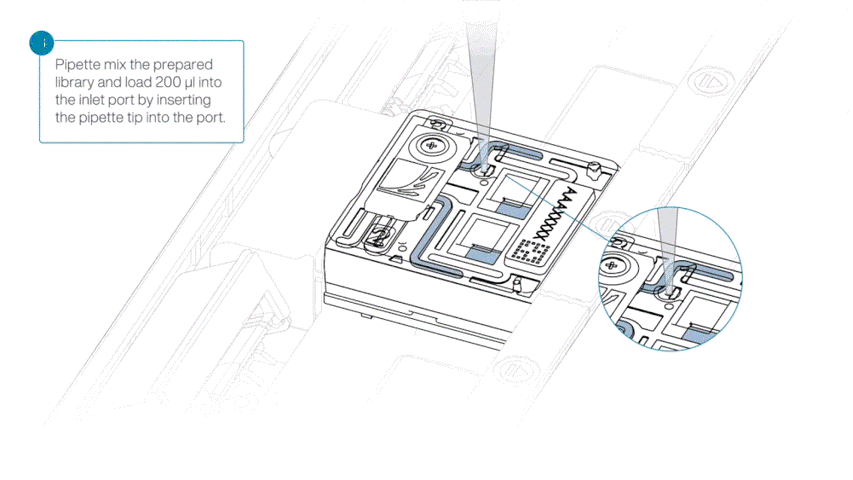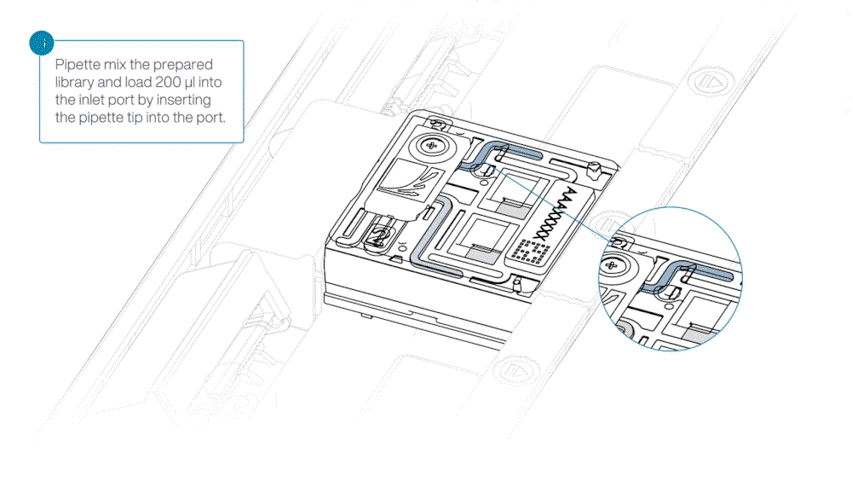
Load 200 µl of library into the inlet port using a P1000 pipette.
Slowly twist the pipette wheel down to load the flow cell (if possible with your pipette) or push down the plunger very slowly.
Note: Dispensing the 200 µl of sequencing library into your flow cell should take at least 5 seconds, ensuring it is done very slowly and at a continuous rate.
Close the valve to seal the inlet port.
Install the light shield on your flow cell as soon as library has been loaded for optimal sequencing output.
We recommend leaving the light shield on the flow cell when library is loaded, including during any washing and reloading steps. The shield can be removed when the library has been removed from the flow cell.
If the light shield has been removed from the flow cell, install the light shield as follows:
- Align the inlet port cut out of the light shield with the inlet port cover on the flow cell. The leading edge of the light shield should sit above the flow cell ID.
- Firmly press the light shield around the inlet port cover. The inlet port clip will click into place underneath the inlet port cover.


Close the PromethION lid when ready to start a sequencing run on MinKNOW.
Wait a minimum of 10 minutes after loading the flow cells onto the PromethION before initiating any experiments. This will help to increase the sequencing output.
Library storage recommendations
We recommend storing libraries in Eppendorf DNA LoBind tubes at 4°C for short term storage or repeated use, for example, reloading flow cells between washes. For single use and long-term storage of more than 3 months, we recommend storing libraries at -80°C in Eppendorf DNA LoBind tubes. For further information, please refer to the DNA library stability Know-How document.
8. Option 2: Store your flow cell for later use
材料
- 测序芯片清洗剂盒(EXP-WSH004)或测序芯片清洗试剂盒 XL(EXP-WSH004-XL)
仪器
- P1000 移液枪和枪头
- P20 移液枪和枪头
Storage Buffer (S) can be used to flush flow cells for storage for later use or to check number of available nanopores before loading another library.
Thaw one tube of Storage Buffer (S) at room temperature.
Slide the inlet port cover clockwise to open the inlet port.
After opening the inlet port, check for a small air bubble under the cover. Draw back a small volume to remove any bubbles:
- Set a P1000 pipette to 200 µl
- Insert the tip into the inlet port
- Turn the wheel until the dial shows 220-230 µl, to draw back 20-30 µl, or until you can see a small volume of buffer entering the pipette tip
Note: Take care when drawing back buffer from the flow cell, do not remove more than 20-30 µl. Visually check that there is continuous buffer from the inlet port across the sensor array.
Slowly load 500 µl of the Storage Buffer (S) into the inlet port, as follows:
- Using a P1000 pipette, take 500 µl of the Storage Buffer (S).
- Insert the pipette tip into the inlet port, ensuring there are no bubbles in the tip
- Slowly twist the pipette wheel down to load the flow cell (if possible with your pipette) or push down the plunger very slowly, leaving a small volume of buffer in the pipette tip.
Note: Dispensing the Storage Buffer (S) into your flow cell should take at least 5 seconds, ensuring it is done very slowly and at a continuous rate.
Remove all of the waste buffers from the waste port by using a P1000 pipette.
You should remove the full volume of waste buffer from the waste channel. If required you can dispense the volume aspirated from the waste channel and repeat the process to remove the full volume from the waste channel.
- Close the inlet port.
- Set your P1000 pipette to 1000 µl.
- Insert the P1000 pipette tip into the waste port.
- Slowly aspirate to remove the waste buffer from the waste port.
Note: As the inlet port is closed, no fluid should leave the sensor array area.
The flow cell can now be stored at 2–8°C.
When you wish to reuse the flow cell, remove the flow cell from storage, and allow it to warm to room temperature for ~5 minutes.
After performing a flow cell wash or storing your flow cell, we recommend using running a 'Flow cell check' to check number of available nanopores.
Load your flow cell into the device with Storage Buffer (S) and start a Flow cell check to detect the number of active pores. For more information, please visit the Flow cell check section of our MinKNOW protocol.
After the Flow cell check, prime your flow cell and load the library before starting a new sequencing run.







