Ligation sequencing gDNA - Cas9 enrichment (Q-SQK-CS9109) (Q_CS9_revA_30Jan2025)
GridION: Protocol
V Q_CS9_revA_30Jan2025
FOR RESEARCH USE ONLY
Contents
Introduction to the protocol
Library preparation
- 4. Preparing the Cas9 ribonucleoprotein complexes (RNPs)
- 5. Dephosphorylating genomic DNA
- 6. Cleaving and dA-tailing target DNA
- 7. Adapter ligation
- 8. AMPure XP bead purification
- 9. Priming and loading the MinION Flow Cell
Sequencing and data analysis
1. Overview of the protocol
Introduction to the CRISPR-Cas protocol
The Cas9 Sequencing Kit (Q-SQK-CS9109) uses the CRISPR-Cas family of proteins which can be programmed to cut specific sequences using oligonucleotide-length RNA, facilitating genome editing and targeted sequencing.
In Cas9 targeted sequencing, we can selectively protect and deprotect ends using CRISPR-Cas and enrich for regions of interest from a given genomic background prior to long read nanopore sequencing of the native DNA sample.
This protocol describes how to carry out sequencing of genomic DNA using the Cas9 Sequencing Kit (Q-SQK-CS9109) with enrichment of specific genomic regions using CRISPR-Cas. You will prepare a target gene region by using gene probes, pooling four probes per region in equimolar quantities. Please refer to the supplementary information for more detail.
Figure 1. The biochemical steps used to prepare your DNA library using the Cas9 Sequencing Kit.
- After DNA extraction, 5’ ends are dephosphorylated to reduce ligation of sequencing adapters to non-target strands.
- Cas9 ribonucleoprotein particles (RNPs), with bound crRNA and tracrRNA, are added to the genomic DNA. They then bind and cleave the Region of Interest (ROI).
- dsDNA cleavage by Cas9 reveals blunt ends with ligatable 5’ phosphates.
- All DNA in the samples is dA-tailed, which prepares the blunt ends for ligation.
- Sequencing adapters are ligated primarily to Cas9 cut sites, which are both 3’ dA-tailed and 5’ phosphorylated. The library preparation is cleaned up to remove excess adapters using AMPure XP beads and resuspended in Elution Buffer. Non-target molecules are not removed. The subsequent library preparation is added to the flow cell for sequencing.
Compatibility of this protocol
This protocol should only be used in combination with:
- Cas9 Sequencing Kit (Q-SQK-CS9109)
- R9.4.1 (Q-FLO-MIN106) MinION Flow Cells
- Flow Cell Priming Kit (Q-EXP-FLP002)
2. Equipment and consumables
材料
- 5 µg high molecular weight genomic DNA at >210 ng/µl (recommended); 1–10 µg (or 0.1–2 pmol) can be used accordingly
- Cas9 Sequencing Kit (Q-SQK-CS9109)
消耗品
- S. pyogenes Cas9 Alt-R™ crRNAs (resuspended at 100 µM crRNA in TE pH 7.5)
- S. pyogenes Cas9 tracrRNA (e.g., IDT Alt-R™, Cat # 1072532, 1072533 or 1072534) resuspended at 100 µM in TE pH 7.5
- Alt-R® S. pyogenes HiFi Cas9 nuclease V3, 100 µg or 500 µg (IDT, Cat # 1081060 or # 1081061)
- Nuclease-Free Duplex Buffer (IDT Cat # 11-01-03-01)
- 1.5 ml Eppendorf DNA LoBind tubes
- 0.2 ml 薄壁のPCRチューブ
- Nuclease-free water (e.g. ThermoFisher, AM9937)
- Agencourt AMPure XP beads (Beckman Coulter, A63881)
装置
- 小型遠心機
- マグネットラック
- ボルテックスミキサー
- サーマルサイクラー
- P1000 ピペット及びチップ
- P200 ピペットとチップ
- P100 ピペットとチップ
- P20 ピペットとチップ
- P10 ピペットとチップ
- P2 ピペットとチップ
- アイスバケツ(氷入り)
- タイマー
オプション装置
- Agilent Bioanalyzer (or equivalent)
- Qubit蛍光光度計(またはQCチェックのための同等品)
- Eppendorf 5424 centrifuge (or equivalent)
For this protocol, you will need 1–10 µg or 0.1–2 pmol high molecular weight genomic DNA (5 µg recommended at a concentration of >210 ng/μl ).
Cas9 Sequencing Kit contents (Q-SQK-CS9109):
| Name | Abbreviation | Cap colour | No. of vials | Fill volume per vial (μl) |
|---|---|---|---|---|
| Adapter Mix | AMX | Green | 2 | 40 |
| Ligation Buffer | LNB | White | 2 | 200 |
| L fragment Buffer | LFB | Orange | 2 | 1,800 |
| S fragment Buffer | SFB | Clear | 2 | 1,800 |
| Loading Beads | LB | Pink | 1 | 360 |
| Elution Buffer | EB | Black | 1 | 200 |
| Sequencing Buffer | SQB | Red | 1 | 300 |
| SPRI Dilution Buffer | SDB | Brown cap, red label | 1 | 1,200 |
| T4 DNA Ligase | LIG | Brown cap, blue label | 1 | 140 |
| dATP | dATP | Brown cap, grey label | 1 | 15 |
| Reaction Buffer | RB | Brown cap, orange label | 1 | 180 |
| Phosphatase | PHOS | Brown cap, yellow label | 1 | 50 |
| TAQ Polymerase | TAQ | Brown cap, green label | 1 | 15 |
| Flush Tether | FLT | White cap, lilac label | 1 | 200 |
| Flush Buffer | FB | Blue | 6 | 1,170 |
Wide-bore tips
Use wide-bore tips (or regular pipette tips with the narrow ends cut off) where possible to minimise shearing of long DNA.
3. Computer requirements and software
GridION IT requirements
The GridION device contains all the hardware required to control up to five sequencing experiments simultaneously and acquire the data. The device is further enhanced with high performance GPU technology for real-time basecalling. Read more in the GridION Q IT Requirements document in the Q system channel.
The sequencing software
The sequencing software controls the GridION, collects sequencing data in real-time and processes it into basecalls. The software can also demultiplex reads by barcode, and basecall/demultiplex data after a sequencing run has completed. You will be using the sequencing software for every assay you run.
For instructions on how to run the sequencing software on the GridION, please refer to the Q-Line sequencing software user guide.
4. Preparing the Cas9 ribonucleoprotein complexes (RNPs)
材料
- Reaction Buffer (RB)
消耗品
- S. pyogenes Cas9 Alt-R™ crRNAs (resuspended at 100 µM crRNA in TE pH 7.5)
- Alt-R® S. pyogenes HiFi Cas9 nuclease V3, 100 µg or 500 µg (IDT, Cat # 1081060 or # 1081061)
- S. pyogenes Cas9 tracrRNA (e.g., IDT Alt-R™, Cat # 1072532, 1072533 or 1072534) resuspended at 100 µM in TE pH 7.5
- Nuclease-Free Duplex Buffer (IDT Cat # 11-01-03-01)
- 0.2 ml 薄壁のPCRチューブ
- 1.5 ml Eppendorf DNA LoBind tubes
- Nuclease-free water (e.g. ThermoFisher, AM9937)
装置
- サーマルサイクラー
- Ice bucket with wet ice
- P200 ピペットとチップ
- P10 ピペットとチップ
- P2 ピペットとチップ
Here, the Cas9 is loaded with crRNA and tracrRNA to form ribonucleoprotein complexes (RNPs) in preparation for the cleavage reaction.
Pre-heat a thermal cycler to 95ºC.
Thaw an aliquot of Reaction Buffer (RB), mix by vortexing, and place on ice.
In an 1.5 ml Eppendorf DNA LoBind tube, pool the crRNA probes for each cleavage reaction by combining equal volumes of each crRNA probe, resuspended at 100 µM in TE (pH 7.5).
- A single crRNA or many crRNA probes may be used in a single cleavage reaction. You will need to validate any use of multiple probes.
- The crRNA probes may also be pre-mixed as an off-catalogue request from IDT.
- For example, you can use probes for the HTT gene, found here, as an individual experiment or in addition to other probes as an in-run control.
- Store any unused crRNA probe mix at -80ºC with minimal freeze-thawing.
Anneal the pooled crRNAs with tracrRNA in Nuclease-Free Duplex Buffer by assembling the following in a 0.2 ml thin-walled PCR tube, as follows:
| Reagent | Volume |
|---|---|
| Nuclease-Free Duplex Buffer (IDT) | 8 µl |
| crRNA pool (100 µM, equimolar) | 1 µl |
| tracrRNA (100 µM) | 1 µl |
| Total | 10 µl |
Mix gently by pipetting and spin down.
Heat the above reaction mix in a thermal cycler at 95ºC for 5 mins.
We do not recommend storing and reusing the annealed mix.
Remove the tube from the thermal cycler and allow it to cool to room temperature (18°C–23°C).
Spin down the tube to collect any liquid in the bottom.
To form Cas9 RNPs, assemble the components in the table in an 1.5 ml Eppendorf DNA LoBind tube in the stated order:
| Reagent | Volume |
|---|---|
| Nuclease-free water | 79.2 µl |
| Reaction Buffer (RB) | 10 µl |
| Annealed crRNA•tracrRNA pool (10 µM) | 10 µl (Step 4, above) |
| HiFi Cas9 (62 µM) | 0.8 µl |
| Total | 100 µl |
The above step yields an excess amount of RNPs, but 10 µl are carried forwards for each reaction into the next target cleavage step. Store any excess RNPs at 4ºC for up to a week.
Thoroughly mix the reaction by gently pipetting and briefly spin down.
Form the RNPs by incubating the tube at room temperature (18°C–23°C) for 30 minutes, then return the RNPs to ice until required.
Proceed to the next step (gDNA dephosphorylation) during the 30 min RNP incubation step.
5. Dephosphorylating genomic DNA
材料
- 5 µg high molecular weight genomic DNA at >210 ng/µl (recommended); 1–10 µg (or 0.1–2 pmol) can be used accordingly
- Phosphatase (PHOS)
- Reaction Buffer (RB)
消耗品
- 1.5 ml Eppendorf DNA LoBind tubes
- Nuclease-free water (e.g. ThermoFisher, AM9937)
- 0.2 ml 薄壁のPCRチューブ
装置
- サーマルサイクラー
- P100 ピペットとチップ
- P10 ピペットとチップ
This step reduces background reads by removing 5’ phosphates from non-target DNA ends.
Prepare the DNA in nuclease-free water.
- Transfer 5 μg genomic DNA into a 0.2 ml thin-walled PCR tube
- Adjust to 24 μl with nuclease-free water
- Mix thoroughly by flicking the tube, avoiding unwanted shearing
- Spin down briefly in a microfuge
Mix the Phosphatase (PHOS) in the tube by pipetting up and down. Ensure that it is at room temperature (18°C–23°C) before use.
Assemble the following components in a clean 0.2 ml thin-walled PCR tube:
| Reagent | Volume |
|---|---|
| Reaction Buffer (RB) | 3 µl |
| HMW genomic DNA (at ≥210 ng/µl) | 24 µl |
| Total | 27 µl |
Note: For an initial test, we recommend a 5 µg genomic DNA input. Target coverage scales linearly with input amount, so the input amount may be reduced accordingly if a lower output is acceptable.
Mix the components by gently flicking the tube and spin down.
Add 3 µl of PHOS to the tube.
Mix the components by gently flicking the tube and spin down.
Incubate in a thermal cycler at 37ºC for 10 minutes, then 80ºC for 2 minutes.
Remove the tube from the thermal cycler and allow to cool to room temperature (18°C–23°C).
6. Cleaving and dA-tailing target DNA
材料
- Dephosphorylated gDNA from the previous step
- crRNA-tracrRNA-Cas9 ribonucleoprotein complexes (RNPs) from the previous step
- Taq Polymerase (TAQ)
- dATP (dATP)
消耗品
- 1.5 ml Eppendorf DNA LoBind tubes
- 0.2 ml 薄壁のPCRチューブ
装置
- サーマルサイクラー
- Ice bucket with wet ice
- ボルテックスミキサー
- P100 ピペットとチップ
- P20 ピペットとチップ
- P10 ピペットとチップ
- P2 ピペットとチップ
In this step, Cas9 RNPs (see 'Preparing the Cas9 ribonucleoprotein complexes') and Taq polymerase are added to the dephosphorylated genomic DNA sample.
This process cleaves target and dA-tails all available DNA ends in one step, activating the Cas9 cut sites for ligation.
Thaw the dATP tube, vortex to mix thoroughly and place on ice.
Spin down and place the tube of Taq Polymerase (TAQ) on ice.
To the PCR tube containing 30 µl dephosphorylated DNA sample, add:
| Reagent | Volume |
|---|---|
| Dephosphorylated genomic DNA sample | 30 µl |
| Cas9 RNPs | 10 µl |
| dATP | 1 µl |
| Taq Polymerase (TAQ) | 1 µl |
| Total | 42 µl |
Carefully mix the contents of the tube by gentle inversion, then spin down and place the tube in the thermal cycler.
Incubate in the thermal cycler at 37ºC for 60 minutes, then 72ºC for 5 minutes, then hold at 4ºC or return the tube to ice.
7. Adapter ligation
材料
- Cleaved and dA-tailed DNA sample
- Ligation Buffer (LNB)
- Adapter Mix (AMX)
- T4 Ligase (LIG)
消耗品
- 1.5 ml Eppendorf DNA LoBind tubes
- Agencourt AMPure XP beads (Beckman Coulter™, A63881)
装置
- Ice bucket with wet ice
- ボルテックスミキサー
- P1000 ピペット及びチップ
- P100 pipette
- P20 ピペットとチップ
Here, AMX adapters from the Cas Sequencing Kit (Q-SQK-CS9109) are ligated to the ends generated by Cas9 cleavage.
Thaw the Ligation Buffer (LNB) at room temperature (18°C–23°C), spin down and mix by pipetting. Due to viscosity, vortexing this buffer is ineffective. Place on ice immediately after thawing and mixing.
Spin down the Adapter Mix (AMX) and place on ice.
Bring the AMPure XP beads to room temperature.
Carefully transfer the contents of the 0.2 ml thin-walled PCR tube to a fresh 1.5 ml Eppendorf DNA LoBind Tube using a wide-bore pipette tip.
Assemble the following at room temperature in a separate 1.5 ml Eppendorf DNA LoBind Tube, adding Adapter Mix (AMX) last, before you are ready to begin the ligation:
| Reagent | Volume |
|---|---|
| Ligation Buffer (LNB) | 20 µl |
| Nuclease-free water | 3 µl |
| T4 Ligase (LIG) | 10 µl |
| Adapter Mix (AMX) | 5 µl |
| Total | 38 µl |
Mix the ligation mix by pipetting up and down slowly and carefully 10 times.
Add 20 µl of the adapter ligation mix to the cleaved and dA-tailed sample. Mix the components slowly and carefully by pipetting. Briefly spin down to collect material at the bottom of the tube.
DNA precipitation
A white precipitate may form upon addition of the adapter ligation mix to the dA-tailed DNA. Adding the ligation mixture in two parts helps to reduce precipitation. However, the presence of a precipitate does not indicate failure of ligation of the sequencing adapter to target molecule ends.
Immediately after mixing, add the remainder of the mix to the cleaved and dA-tailed sample, yielding an 80 µl ligation mix.
Mix the components by gently flicking the tube and spin down.
Incubate the reaction for 10 minutes at room temperature (18-23ºC).
8. AMPure XP bead purification
材料
- Long Fragment Buffer (LFB)
- Elution Buffer (EB)
- SPRI Dilution Buffer (SDB)
消耗品
- Agencourt AMPure XP beads (Beckman Coulter™, A63881)
- 1.5 ml Eppendorf DNA LoBind tubes
装置
- P1000 ピペット及びチップ
- P200 ピペットとチップ
- P20 ピペットとチップ
- 1.5 mlエッペンドルフチューブに最適のマグネット式ラック
- Eppendorf 5424 centrifuge (or equivalent)
This step removes excess unligated adapters and other short DNA fragments, and concentrates and buffer-exchanges the library in preparation for sequencing.
Thaw the Elution Buffer (EB) and SPRI Dilution Buffer (SDB) at room temperature, mix by vortexing, spin down and place on ice.
Thaw one tube of Long Fragment Buffer (LFB) at room temperature and mix by vortexing, then spin down and place on ice.
Add 1 volume (80 µl) of the SPRI Dilution Buffer (SDB) to the ligation mix. Mix gently by flicking the tube.
Resuspend the AMPure XP beads by vortexing.
Add 0.3x volume (48 µl) of AMPure XP Beads to the ligation sample. The volume of beads is calculated based on the volume after the addition of SDB. Mix gently by inversion. If any sample ends up in the lid, spin down the tube very gently, keeping the beads suspended in liquid.
Incubate the sample for 10 minutes at room temperature. Do not agitate or pipette the sample.
Spin down the sample and pellet on a magnet. Keep the tube on the magnet, and pipette off the supernatant when clear and colourless.
Wash the beads by adding 250 μl Long Fragment Buffer (LFB). Flick the beads to resuspend, then return the tube to the magnetic rack and allow the beads to pellet. Remove the supernatant using a pipette and discard.
Repeat the previous step.
Spin down and place the tube back on the magnet. Pipette off any residual supernatant. Allow to dry for ~30 seconds, but do not dry the pellet to the point of cracking.
Remove the tube from the magnetic rack and resuspend pellet in 13 µl Elution Buffer (EB). Incubate for 10 minutes at room temperature.
Note: For longer targets, an extended incubation time may be beneficial.
Pellet the beads on a magnet until the eluate is clear and colourless.
Remove and retain 12 µl of eluate which contains the DNA library in a clean 1.5 ml Eppendorf DNA LoBind tube.
- Dispose of the pelleted beads
The prepared library is used for loading onto the flow cell. Store the library on ice until ready to load.
Library storage recommendations
We recommend storing libraries in Eppendorf DNA LoBind tubes at 4°C for short term storage. For single use and long-term storage of more than 3 months, we recommend storing libraries at -80°C in Eppendorf DNA LoBind tubes.
9. Priming and loading the MinION Flow Cell
材料
- Flush Tether (FLT)
- Flush Buffer (FB)
- Sequencing Buffer (SQB)
- Loading Beads (LB)
消耗品
- 1.5 ml Eppendorf DNA LoBind tubes
- Nuclease-free water (e.g. ThermoFisher, AM9937)
装置
- MinION Flow Cell
- P1000 ピペット及びチップ
- P100 ピペットとチップ
- P20 ピペットとチップ
- P10 ピペットとチップ
Thaw the Sequencing Buffer (SQB), Loading Beads (LB), Flush Tether (FLT) and one tube of Flush Buffer (FB) at room temperature before mixing the reagents by vortexing, and spin down at room temperature.
To prepare the flow cell priming mix, add 30 µl of thawed and mixed Flush Tether (FLT) directly to the tube of thawed and mixed Flush Buffer (FB), and mix by vortexing at room temperature.
Slide open the GridION lid and insert the flow cell.
Press down firmly on the flow cell to ensure correct thermal and electrical contact.
Slide the flow cell priming port cover clockwise to open the priming port.
Take care when drawing back buffer from the flow cell. Do not remove more than 20-30 µl, and make sure that the array of pores are covered by buffer at all times. Introducing air bubbles into the array can irreversibly damage pores.
After opening the priming port, check for a small air bubble under the cover. Draw back a small volume to remove any bubbles:
- Set a P1000 pipette to 200 µl
- Insert the tip into the priming port
- Turn the wheel until the dial shows 220-230 µl, to draw back 20-30 µl, or until you can see a small volume of buffer entering the pipette tip
Note: Visually check that there is continuous buffer from the priming port across the sensor array.
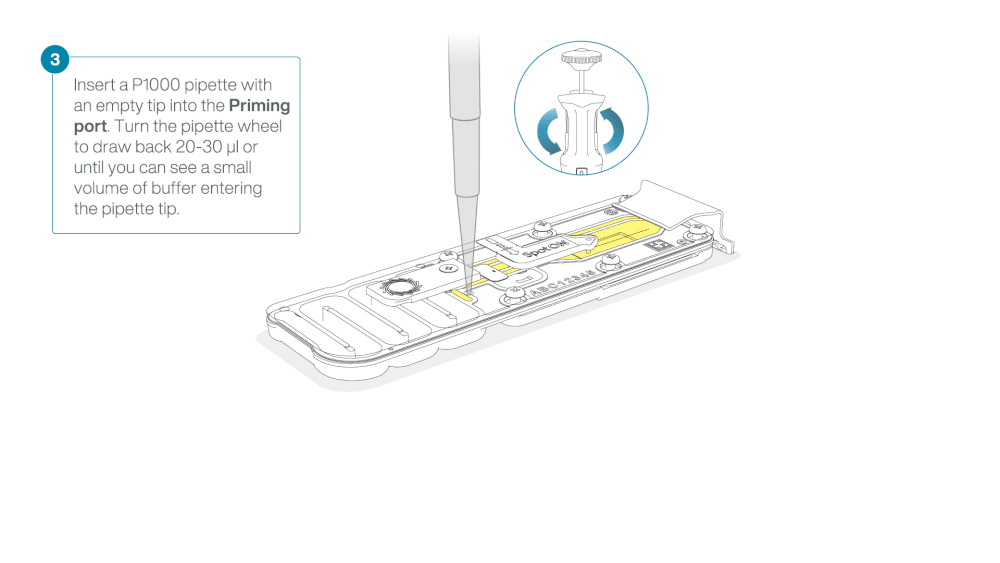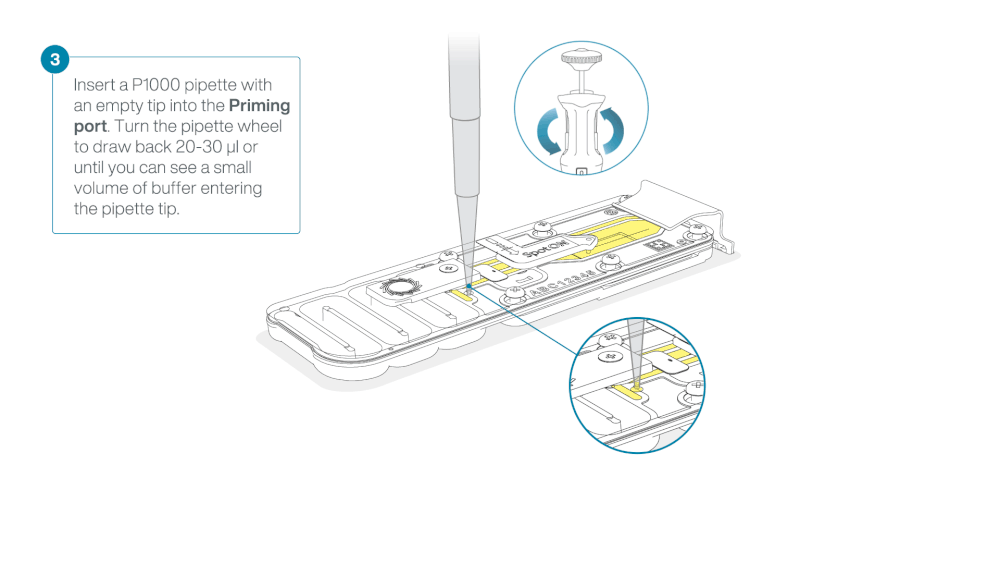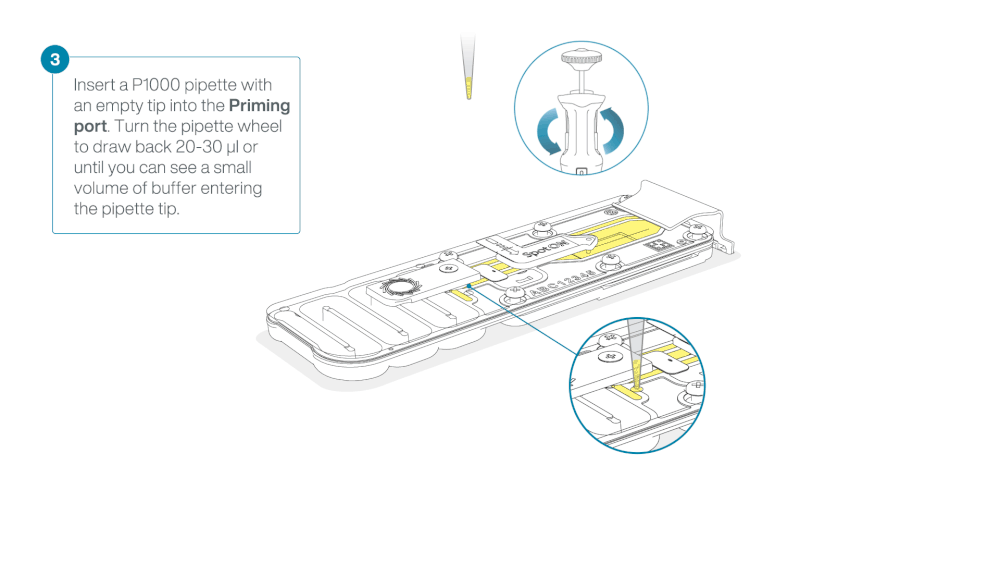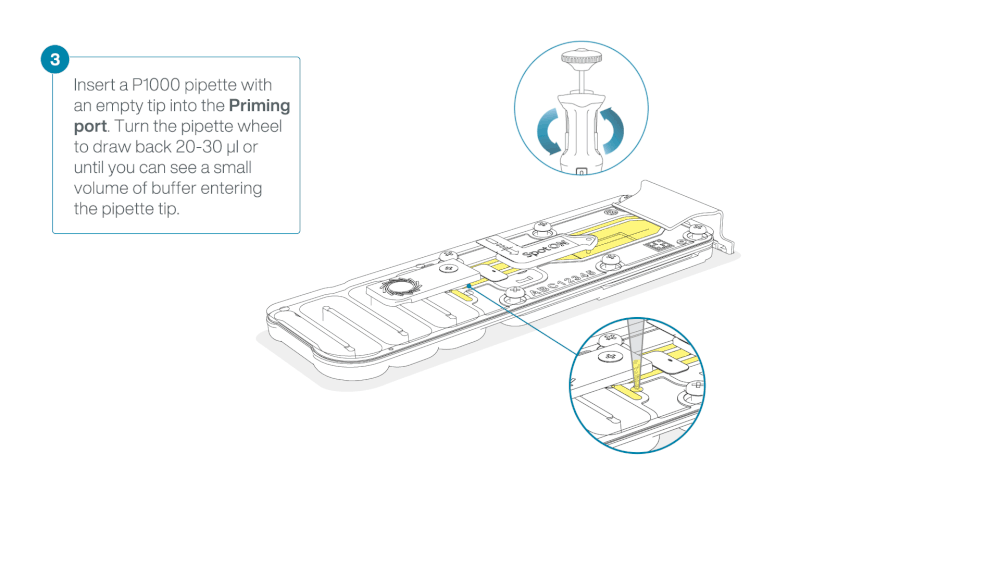
Load 800 µl of the priming mix into the flow cell via the priming port, avoiding the introduction of air bubbles. Wait for five minutes. During this time, prepare the library for loading by following the steps below.

Thoroughly mix the contents of the Loading Beads (LB) by pipetting.
The Loading Beads (LB) tube contains a suspension of beads. These beads settle very quickly. It is vital that they are mixed immediately before use.
In a new tube, prepare the library for loading as follows:
| Reagent | Volume per flow cell |
|---|---|
| Sequencing Buffer (SQB) | 37.5 µl |
| Loading Beads (LB), mixed immediately before use | 25.5 µl |
| DNA library | 12 µl |
| Total | 75 µl |
Note: Load the library onto the flow cell immediately after adding the Sequencing Buffer (SQB) and Loading Beads (LB) because the fuel in the buffer will start to be consumed by the adapter.
Complete the flow cell priming:
- Gently lift the SpotON sample port cover to make the SpotON sample port accessible.
- Load 200 µl of the priming mix into the flow cell priming port (not the SpotON sample port), avoiding the introduction of air bubbles.

Mix the prepared library gently by pipetting up and down just prior to loading.
Add 75 μl of the prepared library to the flow cell via the SpotON sample port in a dropwise fashion. Ensure each drop flows into the port before adding the next.

Gently replace the SpotON sample port cover, making sure the bung enters the SpotON port, close the flow cell priming port and close the GridION lid.
10. Data acquisition and basecalling
How to start sequencing
Follow the instructions in the Q-Line sequencing software user guide beginning from the "Set up and run an assay" section.







