19 highlights from 2019
Dear all,
It’s been another big year for us at Oxford Nanopore – we’ve been thinking a lot about growth. How do we continue to grow the performance of the technology, grow the volume at which we can supply you high quality product, grow the variety of applications that you can do? All this, while preserving our focus on innovation, collaboration, and doing things differently.
However, we don't work on the technology alone. The contribution of the nanopore community, consistently providing feedback, developing tools/methods and supporting each other, is critical, and we thank you.
We are as ever, focused on enabling the analysis of any living thing by anyone, anywhere. We got closer in 2019, but 2020 will be huge. We hope you will enjoy our 19 highlights of 2019.
Gordon
1. The human genome isn't yet complete - but it's getting better
Throughout 2019, the gaps in the human genome have been disappearing. In May, Mark Ebbert’s group (Mayo Clinic) shed light on the "dark and camouflaged genes" of the human genome, newly resolved with long-read nanopore sequencing. We also heard from Karen Miga (UCSC) on how the Telomere-to-Telomere Consortium used long nanopore reads to assemble a complete human X chromosome, and the year closed with Glennis Logsdon (University of Washington), who presented the complete linear assembly of chromosome 8, highlighting that “we are in a very exciting time in genomics”. We expect to see more telomere-to-telomere assemblies in 2020.

2. The long and the short of it
Nanopore is best known for sequencing the longer fragments of DNA/RNA, and the 2018 record still stands at 2.3 Mb. But a nanopore will sequence the fragment that you choose to give it, and this year the conversation also turned towards sequencing shorter fragments. In May, Jeroen de Ridder (UMC Utrecht) presented methods and first clinical trial results using nanopore sequencing to rapidly characterise TP53 from circulating tumour DNA (ctDNA) in head and neck cancer. You can also hear more about short and ultra-short DNA sequencing in this year’s NCM update from the Applications team, featuring their work on ctDNA detection and using short amplicons to detect cystic fibrosis-associated mutations.
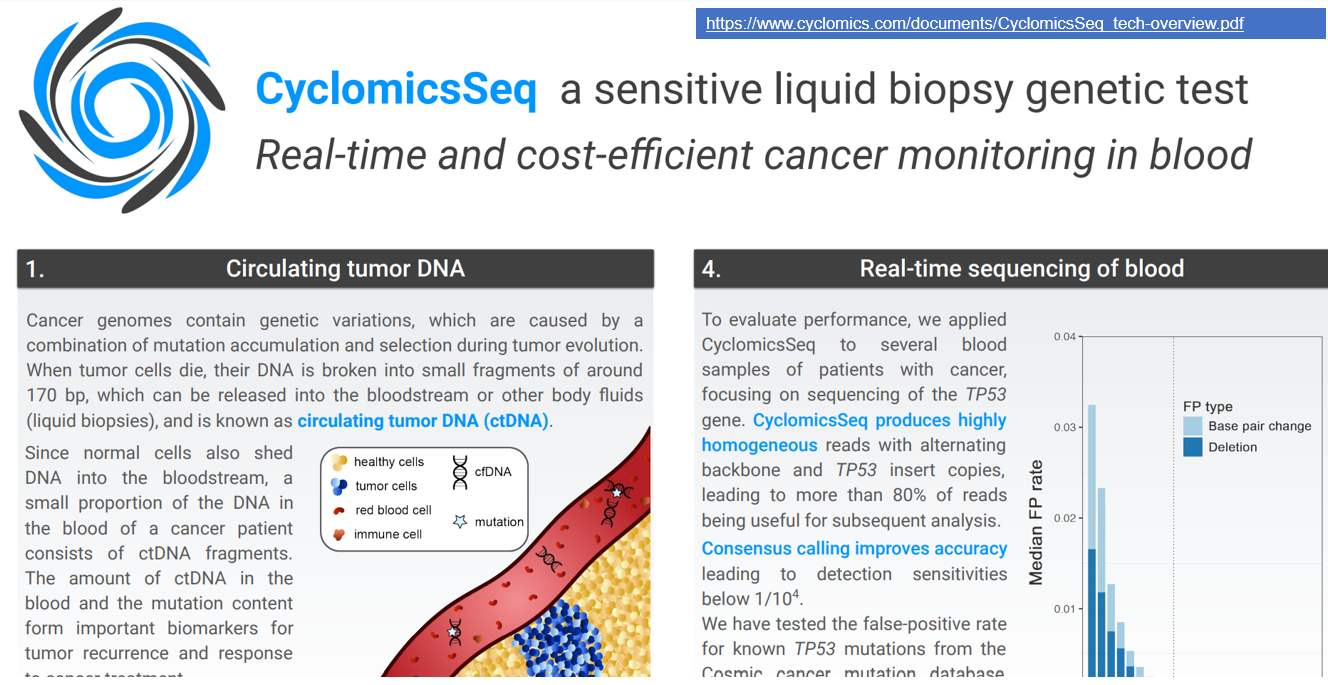
3. Tales of the unexpected
Since the early days of the MinION, the Nanopore Community have been showing us what’s possible with portable, accessible technology – with many surprises along the way. We never could have guessed that in 2019 your experimental subjects would include the upside-down mangrove jellyfish, the Komodo dragon, illegal petunias, the greenfin horse-faced filefish, the polar bear and an ancient Egyptian tomb.

4. Scaling up… and up… and up…
We’ve seen your appetite for nanopore data at large scale evolve from a good meal to a feast through 2019. This corresponded to the release of PromethION 48 in mid-2019, which this year showed 7.3Tb in a single run.
In July, Kishwar Shafin (UCSC) et al. showed how they sequenced 11 human genomes in 9 days on the PromethION. Fast-forwarding to November, deCODE genetics released their work sequencing the genomes of 1,817 Icelanders, the largest ever study involving longer reads. The study revealed a median of 23,111 structural variants per individual, where 2-5,000 had been found with short read sequencing of the same genomes.
As the year closed, the newest population sequencing project was announced by the Department of Health in Abu Dhabi. With an initial phase of 100,000 genomes, the project has chosen to sequence using Oxford Nanopore’s PromethION 48, aiming to deliver the most comprehensive genomes of any population project. We are seeing more and more customers interested in this sequencing at scale with nanopore, and not only on human genomes - sequencing plant genomes at volume is newly enabled with PromethION.

5. Arrival of the littlest flow cell
Sometimes, when only a little data is needed to answer your biological question, even the MinION isn’t quite mini enough. In March, we commercially introduced Flongle: a “Flow Cell Dongle” that adapts the MinION for even smaller flow cells for small-scale, real-time experiments. We were blown away by the surge of demand, and accelerated scaling up of production at the end of 2019. So far, Flongle has been used to demonstrate rapid gene fusion detection, investigate structural variants and methylation in cancer cell lines, genotype Enterovirus, even for low pass CNV analysis of a whole human genome, and more. With Flongle available at less than $100 per flow cell, we’re very excited to see how and where it will be used in 2020.

6. A sustainable nanopore mascot
When we started using WoolCool’s sustainable packaging to keep our kits cool in shipping, we didn’t think it would start a new line in crafts. Over at the Van Tyne lab (University of Pennsylvania), one scientist used the wool packing material to craft this little Christmas mouse.

7. Scaling up production: a new factory – and it looks familiar
In July, our first production processes came online at our brand new high-tech factory in Oxford. Opening an automated factory takes years of planning, as new robotic processes need to be designed and tested, and the team delivered a completed factory ahead of schedule. The building will have capacity for more than a million flow cells a year within four years.
And Gordon asked the team to make the factory look like a MinION, so they did.

8. A nanopore transcriptome from a single cell
Single cell analyses have attracted a lot of attention in 2019, and the nanopore community is exploring this too. In November, a team at Université Côte d'Azur showed how nanopore sequencing and UMIs can be used for high-accuracy isoform detection in the transcriptomes of single cells. On the same theme, Roger Volden (UCSC) explained how he and his team achieved high single-molecule accuracy for multiplexed single-cell cDNA sequencing.
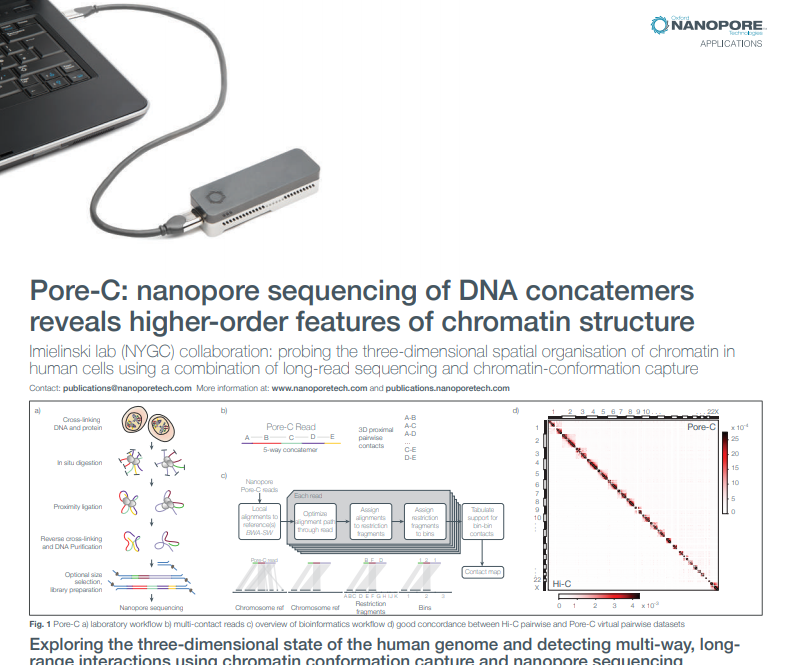
9. 3D DNA
It’s not just the linear sequence of DNA that’s biologically significant - it’s also how the DNA interacts in 3D space. In November, the Oxford Nanopore Applications team released Pore-C: a new method that reveals this, by combining chromatin conformation capture with long nanopore reads. The technique can also be used to dramatically improve assemblies. Pore-C was developed in collaboration with the Imielinski Lab (Weill Cornell); read all about it in the pre-print.
10. And now for something completely different: #TetrisChallenge
Ever wondered what ingredients make up a nanopore sequencing lab? Daphne Hoh (Sen-Lin Tang Microbial Lab) and team showed us how, with their lab’s Tetris Challenge – GridION, pipettes, scuba diver and all.
My lab’s #TetrisChallenge ??????We perform sampling from mountain ⛰ to the sea ? exploring biodiversity ? and their genomic features ? using @nanopore pic.twitter.com/kl4mXh0DLK
— Daphne Hoh (@DaphneHohZW) September 26, 2019
11. Straight to the target - CRISPR/Cas9 gets stuck into nanopore sequencing
When sequencing an entire genome is impractical, picking out DNA of interest to sequence to greater coverage can save on time and costs. This year, researchers have been using PCR-free CRISPR/Cas9 target enrichment to do just that - without sacrificing read length. The method enables long targets to be cut out of a larger sample and sequenced. It has been used by Christina Stangl (UMC Utrecht) and team for accurate, unbiased detection of fusion genes, by Shruti Iyer (Cold Spring Harbor Laboratory & Stony Brook University) to sequence BRCA1 end-to-end in a single read, and lots more.

12. Anything, anyone, anywhere
The Nanopore Community has been sequencing in an ever-growing list of places around the world through 2019, including in low-resource settings and outside of the limits of the laboratory.
The integration of the technology with local infrastructure has also matured. One of the first field applications for nanopore was in the 2015 Ebola outbreak; this year, following the latest outbreak of Ebola, the ARTIC network worked with local organisations in the surveillance of the virus in the Democratic Republic of the Congo.
Charles Kayuki and Laura Boykin of the Cassava Virus Action project showed how nanopore sequencing is helping farmers in East Africa fight crop diseases, whilst MARPLE: a mobile lab for the detection of plant fungal pathogens, was developed for point-of-care plant disease surveillance. At NCM, we heard from Louise Cerdeira (University of São Paulo), who has developed a low-cost, solar-powered analysis unit for long nanopore reads.
When considering 'anyone', we try to make workflows as easy as possible, and price for the lowest possible access requirements.
We plan for easier and automated upstream processes, such as easy sample prep with VolTRAX - which was, for example, used to prepare samples from permafrost in 2019.
However, we also need to think about downstream processes; making sure that biologists without extensive bioinformatics expertise can gain useful results from nanopore sequencing. Beyond that ultimately we would like people without a technical background to be able to analyse anything, anywhere. To address the bioinformatics point, this year we made available extensive training materials to support analysis of nanopore data. These complement the protocol builder, to make sure that you can use the right workflow for your experiment.
Having a great time with @PlacideMbala and team doing a workshop on virus genome sequencing at @INRB_Kinshasa - here looking at full-length EBOV genomes using @NetworkArtic RAMPART. Can you spot @ig299 in this photo? pic.twitter.com/u8echEVTZN
— Nick Loman (@pathogenomenick) December 12, 2019
Some throw back pics of us with our gadgets. 2020 will be epic.
— Dr. Laura Boykin (@laura_boykin) December 7, 2019
#sequencing4farmers pic.twitter.com/FmjPYJVJHE
13. How do I keep flow cells warm when I’m on a polar ice cap?
Look no further than this masterclass from Glen Gowers. Glen and his team made use of the MinION and Field Kit for on-site analysis of microbes on the Vatnajökull ice cap, transporting everything they needed by ski and sledge. Read about the truly off-grid expedition in their Sledge Report.
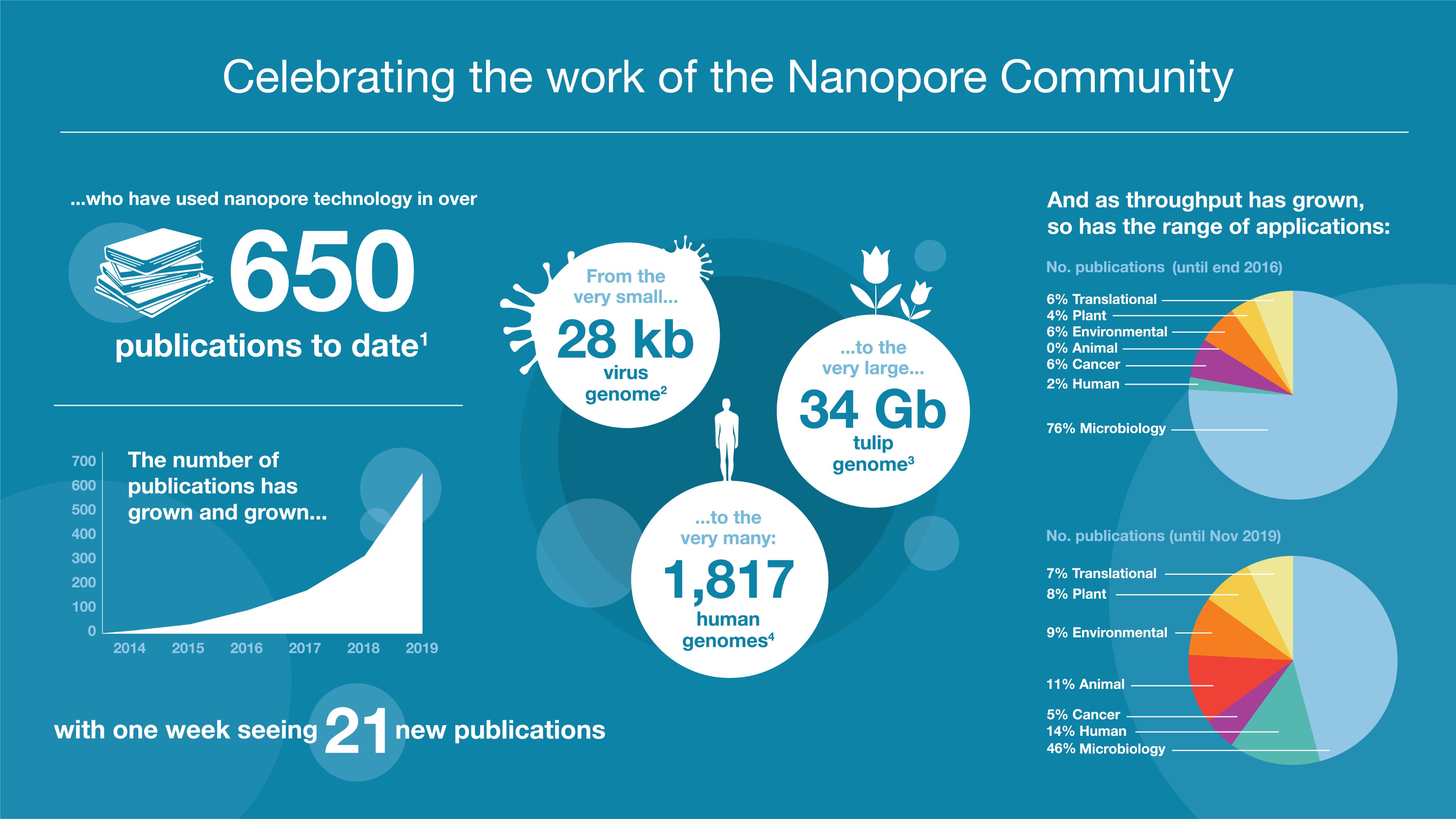
14. More, and more, and more of your publications
Congratulations to everyone whose work has been published this year: more than 650 publications have now featured nanopore sequencing, peaking at 21 papers in a single week this November. The breadth of your work continues to grow; take a look at the Resource Centre for the latest publications from the Nanopore Community.
15. Going beyond canonical bases
Sequencing native DNA (or RNA) with nanopore means going beyond A, T (or U), C and G. In July, methylation detection was integrated into nanopore basecalling, revealing modifications alongside DNA sequence with no additional prep. Watch out for RNA modification calling in 2020, along with extending our capabilities for DNA modifications.
16. A very large tree and a record-breaking scaffold
Assembling the mega genome of a mega tree means breaking some sequencing records. In his talk at Nanopore Community meeting this year, Steven Salzberg (Johns Hopkins University) spoke about how he and his team had assembled the 8.2 Gb giant sequoia genome with long nanopore reads. The assembly included a massive 985 Mb scaffold: “that’s the biggest scaffold ever assembled for any genome – and nobody can break that record unless they get a bigger genome.” Sounds like a challenge for 2020.

17. The most connected sequencer: MinION Mk1C
If you are planning on sequencing anywhere, then it’s useful to have everything you need – a screen, compute and sequencer, in one connected device. In November, we released the first MinION Mk1C instruments to early users. With the first arrivals came unboxing videos from Reindert Nijland and Alexander Wittenberg, then came the sequencing – with Reindert going from unboxing to sequencing in a mere four hours. We're excited to see how you use your Mk1C in 2020 (order yours here!)
#unboxing video of the Mk1C arriving @WUR @nanopore pic.twitter.com/vfJkmPMV6u
— Reindert Nijland (@ReindertN) November 8, 2019
18. Towards the clinic: accurate tests, but faster, and more information
One of the ways we work hard on continuous improvement of the quality of Oxford Nanopore data is to release new data analysis methods, whether new basecallers or other analysis tools. Focusing on high-quality variant calling, this year the R&D team released Medaka to support higher and higher SNV-calling performance, and new workflows that support high quality SV analysis.
It's very exciting to see the evolution of the performance reflected in new methods in biomedical applications. Bringing gold standard-matching (or even -beating) assays ever closer to the patient has become business as usual for clinical developers in the nanopore community this year. Detecting the complete range of gene fusions, SNVs, CNVs, indels and SVs that cause leukaemia, within the 3 days critical for treatment selection, is now a reality thanks to talented teams of nanopore users from across the globe and the power of their MinIONs.
The diagnostic chronic lymphocytic leukaemia genome by nanopore sequencing
At ASH, Researchers Discuss Rapid AML Nanopore Sequencing Assay for Analyzing Actionable Alterations
Not to be outdone by the oncologists, game-changing papers have come out of the infectious disease groups in the Nanopore community, kickstarting a new paradigm in same-day tests and AMR detection. These have focused on potential infections in preterm babies, lower respiratory tract infections, and just before 2019 closed - rapid meningitis characterisation in Vietnam. We’re looking forward to speaking more about our new Q line, that will particularly suit people in regulated environments, in 2020!
19.... we want to say thank you for your support at the end of an exciting and very busy year!
We hope you have a good rest, because we’re going to keep you all busy in 2020!






