Q system MinKNOW user guide (Q_MKN_revB_24Feb2023)
Protocol
V Q_MKN_revB_24Feb2023
FOR RESEARCH USE ONLY
Contents
Introduction to the Q-Line sequencing software
Connecting to device
The Graphical User Interface (GUI)
- 5. Homepage
- 6. Hardware check
- 7. Flow cell check
- 8. Experiments page
- 9. Sample sheet upload
- 10. MiniMap index generation
- 11. Application settings
- 12. Host settings
Host settings
- 13. Overview
- 14. Shutting down
- 15. Rebooting
- 16. Data management
- 17. Mount Network Drive
- 18. Tutorials
- 19. Troubleshooting
Starting a sequencing run
Monitoring and troubleshooting your sequencing run
- 21. Refuelling your flow cell
- 22. Checks and monitoring
- 23. Troubleshooting your run from the pore activity plots
Completing a MinKNOW run
Post-run analysis
1. Introduction
Introduction to the sequencing software
The sequencing software carries out several core tasks: data acquisition, real-time analysis and feedback, basecalling, data streaming, controlling the device, and ensuring that the platform chemistry is performing correctly to run the samples. The sequencing software takes the raw data and converts it into reads by recognition of the distinctive change in current that occurs when a DNA strand enters and leaves the pore. It then basecalls the reads and writes out the data into FASTQ and BAM files.
Computer specification
Please refer to the Q system GridION IT requirements document for details of the IT setup required to run a GridION Q.
2. Login and logout
We recommend users log into the MinKNOW software using their Community credentials.
To log in, you must be connected to the internet.
From here, you can also change the language in the user interface in the bottom right corner.
You will be prompted to enter your email address and Nanopore password:
If you experience login issues, please visit the Community Support channel (https://community.nanoporetech.com/support) and use Continue as guest for temporary use.
To log out of the MinKNOW software:
- Click the button in the top right corner, labelled with your initials or 'Guest' and click Logout.

3. Connection Manager
The Connection Manager enables you to connect to your devices.
MinKNOW can be accessed directly on the device or from another computer that remotely connects to the device used for sequencing, and summarises the states of the flow cells in the device.
- Blue tag: direct connection
- Purple tag: remote connection
Click Add host and enter the IP or hostname of a device to add it to the connection manager for remote access.

To interact with a device, click on the device you want to access from the Connection Manager.
You will be navigated to the Sequencing Overview page of the device.
The flow cell state will be displayed here.

You can now navigate through MinKNOW, start a new sequencing experiment, perform post-run analysis, or view previously run experiments on the device.
4. Device security settings
Default security settings
Default security settings for devices requires users to have a Nanopore account to remotely connect to a device.
To change the remote access availability, settings can be updated when directly connected to a device.
Open 'Host settings' when directly connected to a device.
Navigate to the 'Device settings'. Security settings options are located in 'Remote access availability'.
Select options for local and remote access.

5. Homepage
Homepage overview
The MinKNOW Homepage enables you to navigate to:
a. The Start homepage b. Sequencing Overview of connected flow cells c. Recent and current Experiments d. System Messages e. Host Settings f. Connection Manager to connect with other available devices g. Start Sequencing experiment h. Post-run Analysis i. Flow Cell Check j. Hardware Check k. More includes option to generate .mmi from .fasta file or to import a sample sheet l. Guest/initials to logout

Sequencing overview
This page displays the inserted flow cell state and progress of a current sequencing experiment, including pausing, pore scan and basecalling.
Flow cells with no checks:

Sequencing states:
- Pore scanning:

Basecalling catch up:

Run paused:

During a sequencing run, Flow cell health will be displayed from after the first pore scan.

Select the flow cell to open the quick view of the current experiment.

Experiments page overview
The experiments page displays summary information for all experiments and device check data. You can control runs and see real-time information from sequencing flow cells.
All previous runs can be viewed here by altering the number of days available in "Experiments active in the last ... days".

System Messages
All device reports and messages are displayed here.

6. Hardware check
Hardware check
A hardware check must be performed on all new devices or when software has been upgraded. This uses the Configuration Test Cell(s) (CTC), which comes pre-inserted into your device in place of flow cells.
Insert a Configuration Test Cell (CTC) into the device.
Navigate to the start homepage and select the 'Hardware Check' option.

Select 'Start' for the check to begin.

You will be automatically navigated to the Sequencing Overview page.
A loading bar will be displayed under the flow cell during the checks.
The hardware check will complete after approximately one minute.
A hardware check pass is indicated by a green check icon in the top left corner of the flow cell card.
A fail is indicated by an orange check icon.

If the hardware check fails, remove and reinsert the CTC, and run a hardware check again. If the check fails for a second time, please contact Technical Support via email (support@nanoporetech.com) or via LiveChat in the Nanopore Community.
7. Flow cell check
Flow cell check
A flow cell check must be carried out before loading a DNA or RNA library to assess the number of pores available.
Oxford Nanopore Technologies will replace any flow cell that falls below the warranty number of active pores within 12 weeks of purchase, provided the result is reported within two days of performing the flow cell check and the storage recommendations have been followed.
The minimum number of active pores covered by warranty is 800 for GridION Flow Cells.
Navigate to the start homepage and select 'Flow Cell Check'.

When you see the flow cell type and flow cell IDs recognised, click 'Start' to begin.

You will be automatically navigated to the Sequencing Overview page.
A loading bar will be displayed under the flow cell during the checks.
Flow cell health indicators
The quality of the flow cell will be shown as one of the three outcomes on the Sequencing Overview page:
- Yellow exclamation mark: The number of sequencing pores is below warranty.

- Green tick: The number of sequencing pores is above warranty and ready for sequencing.

- Question mark: A flow cell check has not been run on the flow cell during this MinKNOW session.

Note: the indicator of quality will only remain visible during the MinKNOW session when testing occurred. Once the sequencing device has been re-started, the status of the flow cell will be erased.
8. Experiments page
Experiments page
The experiments page displays summary information for all sequencing flow cells and device checks carried out on the device.
Previous runs can be viewed here until the MinKNOW service is restarted (e.g. after a device reboot).
From this page, you can control specific runs and identify real-time information, including flow cell health and reads.
- Run statistics: The total number of reads, estimated and basecalled bases across an experiment, and number of active and total runs
- Run time: The duration of the experiment
- Run state: The current state of the sequencing run: 'Active', 'Basecalling', 'Complete', 'Stopped with error'
- Health: The current flow cell health
The white panel displays a summary of sequencing experiments and the blue panel displays status information of a specific run.

For more status information of a specific run, click on the run to open the quick view, including current temperature and voltage.

Page configuration
Page configuration allows users to choose which graphs to generate in the quick view of an experiment.
To open page configuration, click on a run in the white panel to open the quick view and click on the Default, All, or Custom options to choose which graphs to display.

Tabs:
- Default: Default graphs available
- All: All graphs available
- Custom: Move graphs from 'Available pages' to 'Your pages' to display a custom order of graphs in the GUI. On this page, you can also choose graph order. Click the graph and drag, then click Save.

Graphs
Select Experiment Groups in the top right corner and swipe sideways to view all the graphs presenting data form all previous runs of the same experiment name, including cumulative output for individual and multiple flow cells.
Use the quick view to display information and graphs for specific runs. To open, select a specific run in the white panel and use the arrows to navigate between the graphs.

For more information about the graphs, refer to Check and monitoring in the 'Monitoring and troubleshooting your sequencing run' section.
Pausing
You can pause a single flow cell in a particular position (a run) or every flow cell (an experiment).
Pausing works by dropping the voltage potential over the membrane to 0 mV to maintain a safe environment to add components, such as fuel, more DNA/RNA library or nucleases for a flow cell wash. Data acquisition will continue during this period, but because no DNA is going through pores at 0 mV, no sequencing data will be collected.
Click the yellow Pause button to open a dialogue box and select which flow cell(s) to pause, then click Pause. 
Triggering a pore scan
The pore scan is used to assess the quality of the four wells in each channel to select the best performing pores. A new pore scan can be triggered every time a sequencing experiment is resumed after a pause (e.g. for a flow cell wash), or if the number of sequencing pores has significantly dropped during an experiment.
Navigate to the Experiments page, open a specific run, and click Start pore scan in the run controls. Confirm the flow cell to be scanned and click Start pore scan.

9. Sample sheet upload
For GridION experiments, it may be preferred to upload sample names and corresponding flow cell positions from a CSV file, rather than manually.
Sample sheet example:
FA026858,SQK-RBK110.96,barcoding_run,sequencing_20200522,barcode01,sample_id_5,test_sample
The sample sheet may describe flow cells being run at one or more than one position. The columns available are as follows, though some are optional:
| Column title | Description | Notes |
|---|---|---|
| flow_cell_id | Defines the flow cell ID which applies to the sample sheet row. | Used to identify which positions to apply values to in MinKNOW. This is optional if position_id is specified |
| position_id | Defines the flow cell position which applies to the sample sheet row. | Used to identify which position to apply values to in MinKNOW. This is optional if flow_cell_id is specified. |
| sample_id | Defines the sample ID to be applied in the run | This is optional. An individual position can only have one sample_id assigned to it when starting a run. |
| experiment_id | Defines the experiment ID to be applied | Each row in the sample sheet must contain the same experiment_id value for the sample sheet to only have a single experiment_id value defined overall. All entries in each row will be validated. |
| flow_cell_product_code | Defines the product code of the flow cell | Used to find the correct sequencing script to start the run |
| kit | Defines the kit and any expansion kits used with the sample | Used to find the correct sequencing script to start the run. The sample sheet must contain only one sequencing kit, but expansion kits are not limited. If expansion kits are additionally defined, they should be separated by a space: e.g. SQK-LSK109 EXP-NBD104 EXP-NBD114 |
For experiments that involve barcoding, an alias is associated with each barcode. Additional columns are available for barcoding runs:
| Column title | Description | Notes |
|---|---|---|
| alias | User-specified string which applies a given label to a specific barcode or barcode pair. | The alias cannot be an existing barcode folder name e.g. 'unclassified', 'classified' and 'mixed' are not allowed. The alias must be between 1 and 40 characters. The haracter set needs to match the following format: /^([0-9a-zA-Z-_] and only contain the following characters: numbers, upper/lower-case letters, dashes, and underscores. |
| type | One of the options: test_sample, positive_control, negative_control, no_template_control | This is optional. |
The sample sheet contains one barcoding arrangement column:
| Column title | Description | Notes |
|---|---|---|
| barcode | The barcode identifier for the row e.g. barcode01 |
Column titles are defined within the first row of the sample sheet. These must be defined in lowercase using the column title values listed above.
Sample sheet validation occurs against the hardware and between rows in the sample sheet to ensure validity. Validation occurs when the sample sheet is loaded. Validation and importing the sample sheet requires all the flow cells to be used in the experiment. Be careful to ensure the correct flow cells and positions are used.
Sequencing parameters using a JSON file (for MinKNOW versions 24.06 and earlier):
Alongside a CSV file containing the sample information, users can also upload a JSON file containing the sequencing parameters for their experiments rather than inputting the information manually.
A JSON file in the correct format, containing all default settings, can be exported from MinKNOW and altered:
Run options tab:
"runLengthHours": 72,(Run length. This must be an integer)"enrichDepleteAdaptiveSamplingEnabled": false,"enrichDepleteAdaptiveSamplingRefFile": null,"adaptiveSamplingChannelStart": 1,"adaptiveSamplingChannelEnd": 512,"enrichDepleteAdaptiveSamplingBedFile": null,"shouldEnrichAdaptiveSamplingRef": true,"barcodeBalancingEnabled": false,"barcodeBalancingCustomBarcodes": false,"barcodeBalancingBarcodeSelection": null,"minReadLength": 200,(Read length selection. This can be 20, 200 or 1000)"activeChannelSelection": true,"muxScanPeriod": 1.5,"groupChangePeriod": 16,"reservedPores": true,
Analysis tab:
"basecallingEnabled": true,(Basecalling can be switched on/off as true/false)"modifiedBasecallingEnabled": false,"barcodingEnabled": true,(Barcoding can be switched on/off as true/false)"basecallModel": "dna_r9.4.1_450bps_hac.cfg",(Basecalling model depends on the flow cell and kit combinations. Make sure that the basecall model is stated as a ".cfg")"modifiedBasecallingContext": "","trimBarcodesEnabled": true,(Trim barcodes can be switched on/off as true/false)"requireBarcodesBothEnds": false,(Requirement to have barcodes on both ends can be switched on/off as true/false)"detectMidStrandBarcodes": false,(Detect mid read barcodes on/off as true/false)"overrideMidBarcodingScore": false,(Override mid barcoding score can be switched on/off as true/false)"overrideBarcodingScore": false,(Override barcode score can be switched on/off as true/false)"minBarcodingScore": 60,(Selection of barcode score. This can be an integer between 40 to 100)"minBarcodingScoreMid": 50,(Selection of mid barcode score. This can be an integer between 40 to 100)"alignmentRefFile": null,"alignmentBedFile": null,
Output tab:
"dataOutputLocationType": 0,"offloadOrOutputLocationNavParams": "path": "/data/.""fastQEnabled": true,(FASTQ files can be switched on/off as true/false)"fastQReadsPerFile": 4000,(Number of reads per file. This is an integer)"fastQDataCompression": true,(FASTQ compression can be switched on/off as true/false)"fast5Enabled": true,(FAST5 files can be switched on/off as true/false)"fast5ReadsPerFile": 4000,(Number of reads per file. This is an integer)"fast5DataCompression": "vbz_compress",(Compression type. This can be"vbz_compress"or"zlib_compress")"selectedRawOutput": "fast5","pod5Enabled": false,"pod5ReadsPerFile": 4000,"bamEnabled": false,"bamWriteMultiple": true,"readFilteringEnabled": true,"readFilteringMinQscore": 9,"readFilteringMinReadlength": null,"readFilteringMaxReadlength": null,"readSplittingEnabled": false,"overrideMinReadSplittingScore": false,"minReadSplittingScore": 58,"bulkFileEnabled": false,"bulkFileRaw": "1-512","bulkFileEvents": "1-512","bulkFileReadTable": "1-512","bulkFileRawEnabled": false,"bulkFileEventsEnabled": true,"bulkFileReadTableEnabled": true
Navigate to the Start page.
Select 'More' and click 'Sample sheet import'.

Select and upload the CSV file (for the sample sheet) or JSON file (for run parameters).

Click 'Start' when the CSV or JSON file has been uploaded. A dialogue box will open to select the parameters for the sequencing run.

Error messages that you may see if you make a mistake in the sample sheet:
- Position defined using
position_iddoes not exist - Position defined using
flow_cell_idcannot be found flow_cell_idandposition_idare used but theflow_cell_iddoes not match the flow cell usedflow_cell_product_codeis used but the value doesn't match the flow cell used- Different
sample_idvalues are assigned against the same position whilst setting up a run flow_cell_product_codeandkitdo not match- Zero or more than one sequencing
kitlisted in kit - Barcode kits in
kitdo not exist as an option on the selected protocol typemust be a valid selection from the list of valid types outlined aboveexperiment_idmust exist within each row and must all have the same value
10. MiniMap index generation
Generating a MiniMap index file prior to starting a run
When loading a FASTA file into MinKNOW for alignment, MinKNOW will generate a MiniMap index file as a first step. To save time when setting up a sequencing run with live alignment, you can create a MiniMap index from a FASTA file from the MinKNOW Start page, prior to set-up.
Navigate to the Start page.
Select 'More' and click 'Create .mmi from .fasta'.

Select an input .fasta file to upload and a folder location for the output .mmi file.

11. Application settings
Application settings contains the tutorials, account/login settings and MinKNOW GUI information.
Navigate to application settings from clicking the initials in the top right corner of the GUI.
The Application settings are available on the Connection manager as well as the device homepage.
Below is an example from the Connection manager: 
Tutorials
Tutorials are available here; they are also presented when logging into MinKNOW for the first time. Users can use Recap to view a specific tutorial or use Reset tutorial state to go through all the tutorials again.


Account settings
Navigate here to logout from MinKNOW.
User email address used for login is displayed here, unless logged in as a guest.

Language
Use Select language to change between English and Mandarin.

12. Host settings
Host Settings
This tab opens the options where users can change device settings.

13. Overview
Features of the Host settings
The Host settings have several features to co-ordinate the device. From this area, users are provided with information on system parameters, such as storage, date/time and IP address.
1. Device Settings: The devices can be shut down and rebooted from this page, and settings for the time and date can be altered. Disk space can be managed and peripherals may also be added and ejected from here. 2. Software: Users are able to download the latest software updates from this page. The option to update will only appear when an update is available. 3. File Manager: Stored data on the device can be managed and transferred. 4. Help: Device logs can be exported using the Export Logs function. Logs and temporary data can also be cleared by using the Clean up function. If troubleshooting of the device is required, a Repair problems function is available as well as a MinKNOW restart option.
Select Back to Main Menu to leave host settings.

Select 'Host Settings' in the side panel to open the options.

Use the side panel to navigate through the options.
Device settings:

Software

File manager:

Help:

14. Shutting down
Navigate to the Device settings and click "Shut down".
Click Shut Down in the pop-up to confirm.

15. Rebooting
To restart MinKNOW on GridION, navigate to 'Help' in the host settings and click "Restart MinKNOW".

16. Data management
File manager
Data can be managed and transferred from the file manager tab on the host settings.
Navigate through the tabs to view the data stored:
- Internal tab: Data stored on the connected sequencing device
- Removable tab: Data stored on a connected removable storage device e.g. USB drive
- Network tab: Data stored on a connected network drive. The network drive must be mounted prior.

To remotely access the device as a shared/network drive:
Navigate to device settings and switch the Share toggle on.

A new window will open to create a password for security.

Select 'Set password', and after a few seconds the share toggle will be active.
Data will now be able to be shared and accessible on different networks.

To stop sharing, click the toggle to turn off.
Note: The password will be removed and must be reset when sharing is turned back on.
To manage data between the internal, removable and network tabs:
Switch the Select toggle on to open the greyed out options Copy, Move, Delete or Rename in the bottom right corner.

Select a folder or file to use the options in the bottom right corner.

To use Move and Delete, select a file and choose an option to either move or delete.
Select the file to copy and click 'Copy'.
A file or folder can be copied to one of your drives on the computer connected for further analysis.

Click either Copy or Move to open a new dialogue box with a list of all the drives accessible to the device.
Note: a USB drive or SD card plugged in will appear under the Devices folder.

Click on a drive to open and navigate through the file directory to choose destination. Click either Copy or Move to confirm action.
Navigate through the file directory to choose destination and select copy to paste the data in your chosen destination.
New directories can be created from the device GUI using the New Directory icon.
Click the icon in the bottom right corner to open Create New Directory and type in a directory name. Click Create.

Disk management
Navigate to Device Settings to view available disk space on the device. Any peripherals plugged in, including USB, can be ejected using the Eject option.
A network drive can also be connected to the device by selecting Add Network Drive and filling in the required credentials for connecting using a Samba (SMB) server or a Network File System (NFS).

17. Mount Network Drive
Network drives
Network drives for data storage can be mounted from either the Device Settings or the File Manager in the Network tab:
Device Settings:

File Manager:

Click "Add network drive".
A modal will open. Click either SMB (Samba) or NFS (Network File System) shares.
Use SMB for connecting to a shared drive on a Windows server, and NFS for Linux. If your shared drive is on a macOS machine or NAS (Network-Attached Storage), you can use either SMB or NFS.
SMB: 
NFS: 
To mount SMB shares:
- Enter the host name or IP address of the SMB share. This is acquired from your network admin.
- Enter the drive on the network host that the user wants to share. Take note to start with a '/'. E.g.
/data. - The 'Mount Point' field will auto-populate with the host name and path to the network share. However, you can customise this. In the file manager, it will appear as:
/data/network/mt-111111-datafor example. - Add a username.
- (Optional) Enter a password.
- (Optional) Enter a domain name.
- Click Mount.

Providing the settings were correct, the mounted drive will be added to the 'Disk Management' overview. The user will be able to:
- View the space information for the drive
- Unmount the network drive
- A link to jump to the mount point inside the file manager
To mount NFS shares:
- Enter the host name or IP address of the NFS share. Obtain this from your network admin.
- Enter the drive on the network host that the user wants to share. Take note to start with a '/'. E.g.
/data. - The 'Mount Point' field will auto-populate with the host name and path to the network share. However, you can customise this. In the file manager, it will appear as:
/data/network/mt-111111-datafor example. - Click Mount.

Providing the settings were correct, the mounted drive will be added to the 'Disk Management' overview. You will be able to:
- View the space information for the drive
- Unmount the network drive
- A link to jump to the mount point inside the file manager
18. Tutorials
Tutorials
On opening the MinKNOW software, the tutorials will start to navigate you through the user interface.
Skip tutorials
Tutorials may be skipped if you are familiar with the user interface.
To skip tutorials, select the three dots and choose to either skip all tutorials or just a section.

Review tutorials
Tutorials can be reviewed again by selecting Reset tutorial state in the Tutorials panel of Application Settings.

19. Troubleshooting
On the Help page of host settings, troubleshooting options are available.
Our device FAQs are located here.
To export logs:
For the PromethION 24/48, PromethION 2i, GridION and PromethION 2 Solo connected to a GridION:
- Click Export in the Export Logs section.
When you have successfully exported the logs, you will be notified of where they are located below the button. The logs will be downloaded as a TGZ file in the logs directory.
You can change the location of the logs by using the file manager, where you can easily retrieve them without needing SSH:
- Click on the file next to Logs exported to: in the Export Logs panel. This will open where the logs are located in the File Manager of MinKNOW.
- Use the control panel in the lower right corner of the File Manager to move the log files.

If the PromethION 2 Solo is connected to a stand-alone computer
Log files for each sequencing experiment can be found in:
Windows: C:\data\logs
macOS: /private/var/log/MinKNOW
Linux: /var/log/minknow
To remove logs and temporary data:
In the "Help" page, click Clean up in the Clean Up Files section and confirm in the in the pop-up.
Note: Run data will not be deleted. Logs and temporary data will be removed.

To repair device issues:
Click Run repair in the troubleshooting section of the Help page and confirm in the pop-up. MinKNOW will attempt to repair the following issues:
- Ensure required directories under "/data" exist and have the correct ownership and permissions
- Ensure required files under "/data" have the correct ownership and permissions
- Ensure the GPU is working at maximum performance
- Ensure that Bluetooth is disabled

20. Starting a sequencing run on GridION
Double-click the MinKNOW icon located on the desktop to open the MinKNOW UI.
![]()
We recommend users to log into the MinKNOW software using their Community credentials.
To log in, you must be connected to the internet.
From here, you can also change the language in the user interface in the bottom right corner.

You will be prompted to enter your email address and Nanopore password:
If you experience login issues, please visit the Community Support channel (https://community.nanoporetech.com/support) and use Continue as guest for temporary use.
Select the sequencing device connected to the computer.

Click 'Start Sequencing' on the Start page to set up the sequencing parameters for your experiment.

When running multiple flow cells simultaneously, it may be preferred to upload sample names and corresponding flow cell positions from a CSV file, rather than manually. To do this, upload a sample sheet from the Start page.
See the "Sample sheet upload" section for instructions.
Type in the experiment name and sample ID and choose flow cell type from the drop down menu.
Ensure the experiment name and sample ID does not contain any personally identifiable information.
Note: If sample ID is not filled in, there will be no sample ID in the folder structure.
Click Continue to Kit Selection to move to the next page.

Save settings
Save settings to reload in following runs as outlined in the Saved and default settings tip.
Select Save settings as template once all settings have been chosen.
Fill in settings name and select Save.

Saved and default settings
To use settings saved from previous runs:
1. Click Load settings from template 2. Choose the saved settings to use 3. Select Load
Saved settings can be deleted from this window by selecting Delete.
Once the saved settings are loaded, you can look through all options and make any changes, if needed, before starting the experiment.
The Saved Settings tip explains how to save settings.
To restore default run settings, select the three dots and click Reload Scripts.

You also have the option to join an existing experiment. After selecting the flow cell position, click Join existing and choose a previous experiment to use the same settings.
Select the kit used from the Kit Selection menu.
If you are running a Control Experiment, check the Control box on the right.
Select Continue to Run Options to choose run parameters.

Skip to final review
To skip setting up the other parameters, click Skip to final review.
You can also return to any previous pages by using the Back to... button.
We recommend keeping the default sequencing parameters unless you have a specific need to change them.

Edit run options of the sequencing run.
The run options tab provides variables for run time and the option to use adaptive sampling.
You can edit any of these settings away from their defaults by using the controls on the user interface.
For more information on adaptive sampling, please see our info sheet:
- Adaptive Sampling info sheet
Select Continue to analysis to proceed to the next section.
Advanced User Options
Active channel selection: Active channel selection refers to a feature where if a pore is in the 'Saturated' or 'Multiple' state, the software instantly switches to a new pore in the group. If a pore is 'Recovering', MinKNOW will attempt to revert the pore back to 'Pore' or 'Sequencing' for ~5 minutes, after which it will select a new pore in the group. This maximises the number of pores sequencing at the start of the experiment.
By default, this option is on but can be switched off. Time between pore scans can also be altered.
Reserved pore: The reserve pore feature prioritises consistency and accuracy over immediacy by reserving wells where voltages have dropped until later in the run, such that other wells can catch up. Switch off this feature to fully front-load sequence data acquisition.

Basecall models
Basecalling models can be selected at two stages in the MinKNOW software:
- Setting up an experiment in MinKNOW - the instructions for selecting a model can be found in the section 'Starting a sequencing run' of this protocol.
- Using post-run basecalling in MinKNOW - the instructions for how to basecall data once an experiment has finished can be found in the section 'Post-run analysis' of this protocol.
Select the High accuracy basecalling model.
Please note than only the High Accuracy (HAC) model is recommended for use with the Q system. While the other unverified (Fast and SUP) models can be used, issues have been observed when using multiple SUP models simultaneously and it is strongly advised you avoid running these alongside important experiments.
Select the output data location, format and filtering options.
The output tab provides variables for data output including file name, location and format.

Data saved as:
Check the name of the output file for the data.
Output location:
Confirm the location for the output data file. Click the file path name to open a pop-up to alter file path or create a new file.
Click Choose location on the pop-up to confirm new file location.
Output location storage capacity:
A bar indicator is displayed below the selected data output file location. This bar will display red when the disk space used reaches more than 80% of the total available disk space. Otherwise, the bar will display green. Below the bar, you can see how much drive space is available.

Output format:
Confirm output file type and file compression using the checkboxes.
Note: Below, reads per file can be changed for both raw reads and basecalled reads, but take note:
- Fewer reads per file: reads become more quickly available. However, too few reads per file means MinKNOW may not keep up with writing out files in real-time.
- More reads per file: reduces the number of files especially for amplicon/cDNA experiments that produce a large number of reads. Some downstream analysis tools may have an upper limit on the uploaded file size.
Basecalled reads: The .FASTQ will be on as default if basecalling in real-time.
Click the cog to open further settings:
- File output frequency can be adjusted for basecalled files. These can be produced every 10 minutes (many small files), hourly (fewer larger files) or at the end of the run (single large file).
- For barcoding runs, basecalled reads are split into files by barcode by default.
- Gzip compression of FASTQ files is on by default of basecalled reads. Note: FASTQ Gzip files may prevent some downstream informatics applications.

Raw reads: Raw reads output can be saved as .POD5 or .FAST5.
- For Kit 14 chemistry and SQK-RNA004, .POD5 is the default file output. The .POD5 output generates files hourly.
- The .FAST5 output file is the default format for our previous and "legacy" chemistry kits. The.FAST5 output produces files for every 4000 reads.
.POD5 is a Nanopore-developed file format which stores Nanopore data in a more accessible way and can be used as an alternative to FAST5 output. This output also writes data faster, uses less compute and has smaller raw data file size.
We recommend keeping the default settings. However, the raw read outup files can be changed by clicking the cog.

Aligned reads: The .BAM output is on as default for sequencing runs using live alignment. This can be observed in the settings by clicking the cog next to Basecalled reads.
Filtering
Filtering options can be used to determine which reads are classed into pass or fail files. These options may also be used to determine which predefined reads, read lengths and Qscore during basecalling can be split out in some live graphs.
Click the cog to open the filtering options pop-up and use the GUI to alter Qscore and to define minimum and maximum readlength for a pass read, if required.


Select Continue to final review to proceed.
Further options:
Select Options in the Output Format panel of the Output tab.
FAST5: The number of basecalls that MinKNOW will write in a single file.
- By default, 4000 reads are written out per file
- The files can be compressed with zlib or VBZ (note: VBZ compression will require a VBZ plugin for compatibility with existing tools. This is optimised for nanopore data, with improved compression over zlib)
FASTQ: The number of files that MinKNOW will write in a single folder.
- By default, 4000 files are written into a single folder
- The files can be uncompressed, or compressed with Gzip to ~55% of original size
BAM: Files containing aligned reads (one BAM file per input FASTQ file).
Users can change reads per file:
- Fewer reads per file: reads become more quickly available. However, too few reads per file means MinKNOW may not keep up with writing out files in real-time.
- More reads per file: reduces the number of files especially for amplicon/cDNA experiments that produce a large number of reads. Some downstream analysis tools may have an upper limit on the uploaded file size.

Chosen options can be saved by selecting Save for future use.
Advanced User Options
Option to turn bulk file saving either on or off.
Note: This will result in much larger file sizes due to additional information about the run, which is used for debugging.

Select 'Start' to run the experiment.
The Review page is an overview of all run options selected.
Edits can be made by selecting the Edit button.
Select Advanced run options to view the extra options selected.

You will be automatically navigated to the Sequencing Overview when sequencing starts.
From here, you can see a progress bar below the flow cell to show the progression of the sequencing script.
In some cases, the device will take a few minutes before starting a sequencing run if an alignment reference file is used. A progress bar will show the progress before sequencing begins.
Flow cell health will be displayed after the first pore scan.

Click on the flow cell to open the quick view and check the number of active pores. The first pore scan should report a similar number of active pores (within 10-15%) to that reported in the flow cell check.
Below are recommended troubleshooting tips if there are unexpected differences in pore numbers:
- If there is a significant reduction in active pores in the first pore scan, restart MinKNOW.
- If the numbers are still significantly different, close down the host computer and reboot.
- When the numbers are similar to those reported at the end of the flow cell check, restart the experiment. There is no need to load any additional library after the restart.
To stop the experiment, click 'Stop' on the experiments page.
This is only necessary if you want to stop your run before the end of the set run time.

In the new window, select either 'Stop sequencing' or 'Stop sequencing and basecalling'.

Note: If you select 'Stop sequencing' any catch-up basecalling will still occur. A progress bar will appear on associated flow cell on the Sequencing Overview tab.
Once sequencing is complete, generate a run report.
A run report containing information about the sequencing run and performance graphs are automatically generated but can be generated manually by clicking Export run report and selecting which experiment to export to html.
For more information about the run report, please see the Run report section of this document.
A pore activity .csv file is also generated for every run.

21. Refuelling your flow cell
Refuelling: general advice
Refuelling is the replenishment of motor fuel in the sequencing experiment through the addition of Flush Buffer (FB) from the Flow Cell Priming Kit (EXP-FLP002). The translocation speed graph in MinKNOW can indicate whether it is necessary to top up fuel.
When to refuel
If the DNA translocation speed drops below 300 bases per second, you may start to see a reduction in quality of data reflected in the Qscore. We therefore recommend topping up the flow cell with fuel, using the Flush Buffer (FB) from the Flow Cell Priming Kit. Please follow the instructions below if you wish to top up the fuel during an experiment.
Please consult the 'Priming and loading the SpotON flow cell' step from the relevant library preparation protocol for instructions on adding solutions to the flow cell.
Refuelling a GridION Flow Cell
Instructions for refuelling a GridION Flow Cell:
Remove one tube of Flush Buffer (FB) from the freezer and thaw by bringing to room temperature
Pause the experiment on the GridION position that is being refuelled:
a. Navigate to Experiments and select the experiment running b. Click Pause c. A dialogue box will open. Choose the flow cell to pause and click Pause to confirm

After opening the priming port, check for a small bubble under the cover. Draw back a small volume to remove the bubble (a few µl):
a. Set a P1000 pipette to 200 µl b. Insert the tip into the priming port c. Turn the wheel until the dial shows 220-230 µl, or until you can see a small volume of buffer entering the pipette tip
Visually check that there is continuous buffer from the priming port across the sensor array.
Complete the flow cell refuelling:
- Load 250 µl of the FB into the flow cell via the priming port (i.e. not the SpotON port), avoiding the introduction of air bubbles
- Close the priming port and replace the GridION lid
- Unpause the experiment on the relevant GridION position by clicking Resume
- (optional) Click Start pore scan and choose your flow cell, to pick a new set of the best channels for the remainder of the sequencing experiment.

The outcome and benefits of refuelling
Translocation speed and Qscore over time
Below is a graph that shows what is expected for translocation speed after the addition of FB to the flow cell in a previous version of MinKNOW.
As the speed drops below 300 bases per second, the Qscore begins to decline for the reads processed through the nanopores at this speed. After refuelling at the 17.5 hour mark, the speed begins to increase and returns to an improved rate (~400 bases per second), which is similar to the speed at the start of the experiment. After the addition of fuel, the quality of the data increases and returns to Qscores equivalent to those seen at the start of the run.

Refuelling multiple times in a run
You can refuel a sequencing run multiple times over an experiment. When you should refuel will depend on when the translocation speed drops below 300 bases per second on the speed graph in the MinKNOW UI.
Warning - overloading the flow cell when refuelling
If you refuel your flow cell multiple times, the waste reservoir of the flow cell will gradually fill up with buffer.
The array and waste reservoir of the GridION flow cell will take the following fluid loads:
- priming mix
- a library
- three refuels
22. Checks and monitoring
During the sequencing experiment, you can check various flow cell health and performance parameters that are shown in the MinKNOW GUI. Each parameter is described in more detail in this section.
Flow cell health
During a sequencing experiment, the MinKNOW Sequencing Overview page shows a flow cell icon with coloured bars. The bars represent the combined health of all pores in a flow cell, and indicate how well the flow cell is performing. The colours are:
- Light green: sequencing
- Dark green: open pore
- Dark blue: pore recovering
- Light blue: pore inactive
This information is identical to the last bar of the pore activity plot (described later).

Experiment summary information
The Experiments page displays summary information for all sequencing flow cells and device checks carried out on the device.
From this page, the user is able to control specific runs and identify real-time information including flow cell health and reads, giving users real-time feedback for sequencing flow cells.
- Run statistics: The total number of reads and bases produced across the experiment
- Basecall statistics: There are two values for basecalled reads:
- Basecalled reads as a percentage of the total reads produced across the experiment. This gives an indication as to the size of the queue for reads to be basecalled
- Total number of reads basecalled across the experiment
- Run time: The duration of the experiment
- Temperature: The heatsink temperature of the selected position, which for sequencing should reach 34°C
- Voltage: The applied potential of the position at that point in time

As the MinKNOW script progresses, you can check the following:
- Number of active pores
- Heatsink temperature
- Development of the read histogram
- Pore occupancy
- Local basecalling report
Channel states
The channel states pannel gives an overview of the states the flow cell pores are in to give the user an idea of how well the sequencing run is performing in real time. A good library will be indicated by a higher proportion of light green channels in "Sequencing" than are in "Pore available". The combination of "Sequencing" and "Pore available" indicates the number of active pores at any point in time. A low proportion of "Sequencing" channels will reduce the output of the run.

Clicking on the Show Detailed button reveals a more detailed array of channel states:
- Sequencing: Pore currently sequencing.
- Pore Available: Pore available for sequencing.
- Adapter: Pore currently sequencing adapter.
- Active feedback: Channel ejecting analyte.
- No pore: No pore detected in channel.
- Multiple: Multiple pores detected. Unavailable for sequencing.
- Unavailable: Pore unavailable for sequencing.
- Unclassified: Pore status unknown.
- Saturated: The channel has switched off due to current levels exceeding hardware limitations.
- Out of range-high: Current is positive but unavailable for sequencing.
- Out of range-low: Current is negative but unavailable for sequencing.
- Zero: Pore currently unavailable for sequencings.
Pore activity plots
The pore activity plot summarises the channel states over time. Each bar shows the sum of all channel activity in a particular amount of time. This time bucket defaults to 1 minute, and scales to 5 minutes automatically after 48 minutes. However, bucket size can be adjusted in the "Bucket size" box in Display Settings.
The graph populates over time, and can be used as a way to assess the quality of your sequencing experiment, and make an early decision whether to continue with the experiment or to stop the run.

To see the more detailed view of channel states, click Show detailed.
- Sequencing: Pore currently sequencing.
- Adapter: Pore currently sequencing adapter.
- Pore available: Pore available for sequencing.
- Unavailable: Pore unavailable for sequencing.
- Active feedback: Channel ejecting analyte.
- No pore: No pore detected in channel.
- Out of range-high: Current is positive but unavailable for sequencing.
- Out of range-low: Current is negative but unavailable for sequencing.
- Multiple: Multiple pores detected. Unavailable for sequencing.
- Saturated: The channel has switched off as current level exceed hardware limitations.
- Zero: Pore currently unavailable for sequencing.
- Channel disabled: Channel is disabled and awaiting another pore scan.
Read length histogram
The cumulative histogram shows reads compared to bases. Use the options below to choose the axis legends:
- Y-axis: Estimated bases or basecalled bases
- X-axis: Read length or read counts
Read count - this shows the number of reads vs read length. This enables you to understand how the read lengths vary in number and size.
Read length - this shows the total number of bases vs the read length.
The N50 value is presented (only for the whole set of passed reads) in the top left corner of the histogram.
 Each histogram's X-axis (read length) can be zoomed in using the scaled bar under the histogram. Use Reset to refocus the zoom bar and histogram for the entire 'passed read' dataset.
Each histogram's X-axis (read length) can be zoomed in using the scaled bar under the histogram. Use Reset to refocus the zoom bar and histogram for the entire 'passed read' dataset.  You can see the number of bases by hovering over the bar in question.
You can see the number of bases by hovering over the bar in question.
Reads that are outliers in terms of length can be removed from the graph by ticking the Hide outliers box below the histogram.
Select Split by read end reason to view split reads and hover over for further information. This is useful for adaptive sampling which is further explained in the Adaptive Sampling info sheet.
- Device changed MUX/Pore scan initiated: The strand ended because there was a scheduled pore scan that interrupted the read.
- Read completed: The strand ended naturally as it passed through the pore.
- Read blocked: The strand ended because it was deemed of low quality and purposefully rejected.
- Adaptive sampling voltage reversal/Adaptive sampling rejection: The strand was rejected by adaptive sampling because it did not align to the target sequence when enriching, or the strand matched to the target sequence when depleting.

Cumulative output
The cumulative output graph shows:
- the number of bases that have been sequenced and basecalled

- the number of reads that have been sequenced and basecalled; and whether the reads have passed of failed the quality filters

Cumulative output of multiple flow cells
If there are multiple flow cells running under the same experiment name, you will see the Experiment view. This gives output information on all assigned flow cells, plus a running cumulative total of bases or reads sequenced.
The cumulative output graph shows the running total number of reads or Gbases sequenced by the multi-flow cell platform like GridION. 
The output generated by each flow cell to make the total cumulative output can be represented by individual output plots.
Both graphs can be switched between in the MinKNOW GUI. Reads, bases (estimated or basecalled), sample ID or flow cell ID can be selected to tailor the output graph, as required.
Temperature and Bias Voltage graph
Temperature vs time graph
The temperature graph gives a real-time representation of the heatsink temperature of the flow cell. If the temperature reading drifts out of the target zone, please consult Technical Services, otherwise the quality of your data may be compromised.

Bias voltage vs time graph
The bias voltage graph provides the running voltage in real-time. MinKNOW will automatically adjust the applied voltage and will naturally drift to lower voltages as the electrochemistry in the flow cell is depleted.
You will notice drops in the voltage at regular intervals and these will correspond to the channel scans that are defaulted to occur every one and a half hours. Here, each channel will be scanned to look for its availability for sequencing. The common voltage is reversed before and after each channel assessment for clearer results. 
Translocation speed and quality score graphs
Note: These graphs are only present if basecalling is turned on.
Translocation speed vs time
The translocation graph gives a real-time representation of the speed at which DNA/RNA strands pass through the pore. If the translocation speed drops below this window, data quality and output may be compromised as strands take longer to move through the pore.

Quality score vs time
The Quality score graph gives a live representation of the median strand Q-score over time. 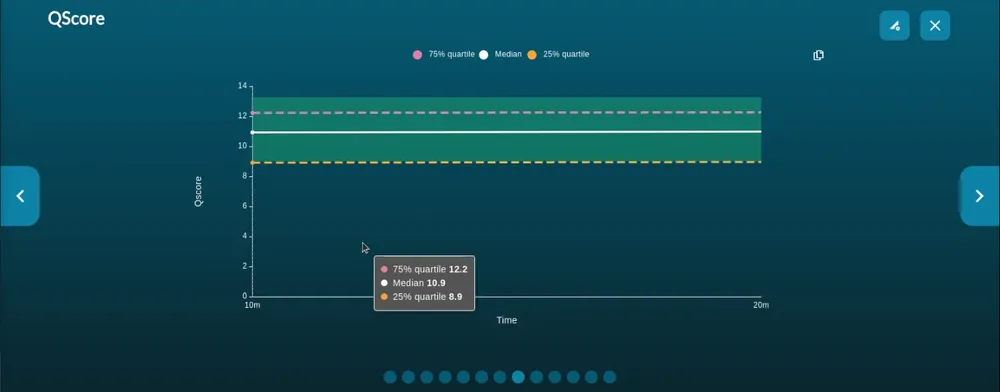
Barcode read counts
The Barcode Read Counts graph shows the breakdown of barcoded reads, if barcoding was used for the experiment. The default view only shows reads that have passed the quality score filters. However, selecting the Display failed box will show all reads. 
The X-axis of the histogram also has a zoom function using the scaled bar underneath. Use Reset to refocus the zoom bar and graph.
Note that there is a small amount of cross-talk between barcodes. Some barcoded reads may appear on the graph even if the barcode in question was not used for the experiment.
Note: This graph is only present if barcoded is turned on.
Alignment hits
The alignment hits graph will populate when alignment and basecalling is set-up to run during sequencing. This bar graph shows the number of reads and bases that align to each of the entries in the user reference .fasta file or minimap index file. An entry in the reference file will only appear on the graph once a single read has aligned to it.

The X-axis of the histogram also has a zoom function using the scaled bar underneath. Use the Reset to refocus the zoom bar and graph.
A target alignment hits view is also available in the same graph by clicking Alignment coverage. This is populated by the .BED targets if a .BED file is supplied with the alignment reference.

Alignment and barcode heatmap
The alignment and barcode heatmap is only available when alignment and demultiplexing is performed with basecalling during sequencing. The heatmap graph shows the alignment hits split per barcode. The colour gradient shows bases and reads which are the more popular barcode and alignment hit combinations. Use the options below to view reads or bases and the slide bar above to focus on specific regions.

Note: These graphs are only present if barcoding and alignment are both turned on.
Traceviewer
The Traceviewer displays the current levels from individual channels. By default, it is set to show 10 channels. This number can be changed through the selection boxes beneath the viewer. Additional parameters that can be altered:
- Time: The length of time plotted on one screen
- Maximum: The highest current level to be shown on the y axis
Please note that viewing a high number of channels in the traceviewer may impact the speed at which the GUI is able to function.
Pore scan
As the sequencing protocol starts, a pore scan begins before the sequencing. There are four groups of active pores, and the pore scan allows MinKNOW to pick the best-performing pores in each group, maximising the data output in the initial stages of the run. The software also instantly switches to a new channel in the group if a channel is in the “Saturated” state, or after ~5 minutes if a channel is “Recovering”.

23. Troubleshooting your run from the pore activity plots
Pore activity plot introduction
The pore activity plot feature in the MinKNOW software can be used to judge the quality of your experiment. The pore activity plot shows the distribution of channel states over time, grouped by time chunks, or 'buckets'. The basic view shows the five main channel states: Sequencing, Pore, Recovering, Inactive, and Unclassified. Clicking the "More" button shows a more detailed breakdown of channel states.
It is recommended to observe the pore activity plot populating over the first 30 min-1 hr of the sequencing run. By this time, the channel state distribution will give an indication whether the DNA/RNA library is of a good quality, and whether the flow cell is performing well.
The software instantly switches to a new channel in the group if a channel is in the “Saturated” or “Multiple” state, or after ~5 minutes if a channel is “Recovering”. This feature maximises the number of channels sequencing at the start of the experiment; however this may also result in an artificially high number of "Sequencing" or "Pore" channels in the pore activity plot. For this reason, we recommend referring to the pore scan plot, which shows the true distribution of channel states at the point of the most recent pore scan.
Good library
A good quality library will result in most of the pores being in the "Sequencing" state, and very few in "Pore", "Recovering" or "Inactive". A library that looks like this is likely to give a good sequencing output.

Channel blocking
Under certain conditions (usually the presence of contaminants in the library), pores may become blocked and therefore unable to sequence. This manifests itself as a build-up of "Unavailable" pores over time.

Recommendation:
- If, despite the channel blocking, the library is still producing a sufficient number reads to answer your biological question, you can carry on with the sequencing experiment.
- Otherwise, stop the sequencing run in MinKNOW. Then wash out the library from the flow cell using the instructions for the Flow Cell Wash Kit. Then prepare another library and load it on the flow cell.
Osmotic imbalance
If the plot shows a high number of "Inactive" channels building up over time, this could indicate that the channels or membranes have been damaged, for example by air bubbles, osmotic imbalance, or the presence of detergents or surfactants in the library.
Pore activity: 
Pore scan: 
Recommendation:
- Check the channel panel: if the Inactive channels are all grouped in one part of the flow cell, this could indicate an air bubble that has been introduced during flow cell flushing or library loading. If the remaining channels are still sequencing, it is possible to carry on with the run. Do not try to move the air bubble, as this can damage even more channels.

If the Inactive channels are distributed throughout the flow cell:
- Check that the heat tape on the underside of the flow cell is intact.
- Make sure that the input DNA is in either TE buffer or nuclease-free water, and that the buffer contains no detergents or surfactants.
- Make a new batch of flow cell priming buffer (a mixture of Flush Buffer/Flow Cell Flush and Flush Tether/Flow Cell Tether). Flush the flow cell with the mixture and load the library again.
Low pore occupancy
If there was insufficient starting material, or some sample has been lost during library prep, or the sequencing adapters did not ligate well to the strand ends, the plot will show a high ratio of "Pore available" to "Sequencing" states, meaning that only a limited number of pores are sequencing at any one time.

Recommendation:
- Check the amount of DNA/RNA in your prepared library, for example by using the Qubit fluorometer. We recommend preparing a fresh library and reloading your flow cell with the recommended loading input from the relevant protocol
- If your library is at a low concentration, prepare the library again using a higher amount of starting material.
- Ensure you are adding the flush tether to your priming mix during flow cell priming and loading before adding your library.
- For ligation-based protocols, ensure you are following the protocol to ligate the sequencing adapter correctly and performing any clean-up steps after adapter ligation with short or long fragment buffer rather than ethanol.
24. Exiting the GUI and shutting down
Shutting down your GridION
The GridION's computer requires a stepwise, processed shut down, otherwise you may face problems e.g. when recognising flow cells.
Follow the instructions in this section to ensure you do not face errors with your GridION from incorrectly shutting down your device.
Ensure none of the flow cells are running an experiment.
 An indication no experiments are running is the lack of an experiment status bar beneath individual flow cells.
An indication no experiments are running is the lack of an experiment status bar beneath individual flow cells.
Navigate to Device Settings and click 'Shutdown'.

Turn off the device at the mains supply, if you are using mains power.
If connecting to a device remotely using a computer not directly connected to the device, close down the MinKNOW application window.
25. Post-run basecalling
Basecalling overview
A user can basecall and demultiplex their data directly in MinKNOW after a sequencing experiment has finished, or re-analyse old data using the latest basecalling models. Reads can also be aligned against a reference post-run.
Note: Both barcoding and alignment can be run on .fastq or .fast5 reads when coupled with basecalling.
Select 'Analysis' on the start page.

Click 'Basecalling' to open the post-run basecalling set-up options.

Select input folder containing the .fast5 files to basecall from a previous run.
Select whether you want to process .fast5 reads in sub-directories.
If your chosen read input folder contains sub-directories with .fast5 files, you can choose whether or not to basecall .fast5 files in these sub-directories by switching this option on.

Select output folder and file type.
By default, MinKNOW will create a /basecalled folder in /data. You can set a different folder in which to save the basecalled reads. Note: This must be a sub-folder of /data directory.
Select whether to output FAST5 files. If selected, these files will be written to a folder within /basecalled called /workspace, if selected.
Users are also able to enable read splitting options post-run.

Choose the basecalling model from the drop-down menu.
The model is used to process raw signal data for a basecall. The model name is composed of:
[flow cell product code] + [sample type] + [model type] e.g. FLO-MIN106 - DNA - HAC

Given the recent improvements to basecalling accuracy, Q-score cut-off for reads put into the "pass" folder has been increased to:
Fast basecalling model >8 High accuracy (HAC) model >9 Super accurate (SUP) model >10
Click 'Start' to begin post-run basecalling.

26. Post-run barcoding
Barcoding overview
A user can barcode their data directly in MinKNOW after a sequencing experiment has finished, using FASTQ data.
Click 'Barcoding' in the analysis menu to open the post-run barcoding set-up options.

Select the input folder containing the FASTQ data to barcode from a previous run.
Select whether you want to process FASTQ reads in sub-directories.
If your chosen read input folder contains sub-directories with FASTQ files, you can choose whether to barcode FASTQ files in these sub-directories by switching this option on.
Note: Data can only be saved to the /data folder.

Choose output folder for the post-run barcoded data.
By default, MinKNOW will create a /barcoding folder in /data/. You can set a different folder which to save the barcoded reads. Note: This must be a sub-folder of /data directory.

Select any barcoding kits used for the run from the drop down menu.
MinKNOW will write out basecalled reads into barcode-specific folders and enable demultiplexing during basecalling.
Users are able to choose specific settings, including trimming barcodes and minimum barcoding scores.

Click 'Start' to begin post-run barcoding.







