PromethION (PRTD_5000_v1_revB_08Aug2018)
TechnicalDocument
PromethION V PRTD_5000_v1_revB_08Aug2018
FOR RESEARCH USE ONLY
Contents
What is it?
How it works
What is needed
Regulatory and Safety
1. An overview of PromethION
An overview of PromethION
PromethION offers the same real-time, long-read, direct DNA and RNA sequencing technology as MinION Mk1B and GridION, at much larger scale enabling users to apply the technology to large projects.

PromethION is a standalone benchtop system designed to run up to 48 flow cells at any time. It consists of a Data Acquisition Unit and a bench-top Sequencing Unit. The Data Acquisition Unit contains the on-board compute, consisting of various processing and memory units to enable mass throughput of data generated by the Sequencing Unit. The Sequencing Unit comprises six sensing modules of eight flow cells.

The system allows on-demand sequencing. Users can start and stop running individual experiments as required, or deploy multiple flow cells onto single experiments for greater speed or throughput. Each PromethION flow cell contains 3000 nanopore measuring channels. Users have achieved in excess of 150 Gb from a single flow cell making the PromethION capable of generating many Terabases of RNA or DNA data in a single run.
PromethION is highly suited to large genome sequencing at scale, whole transcriptome experiments, population studies and projects with very high sample numbers.
The device is simple to install and can be as easily run in a production sequencing environment as in a research lab, enabling different users to start and stop their runs as they require.
This document will describe the PromethION Alpha-Beta Device which is comprised of an advanced, optimised, Data Acquisition Unit and a high-throughput, adaptable Sequencing Unit, currently enabling 24 flow cells to be run simultaneously or individually.
The PromethION is For Research Use Only.
2. The hardware
The Data Acquisition Unit
The Data Acquisition Unit supports the Sequencing Unit by providing the necessary processing and temporary memory - namely 384 GB RAM, to cater for real-time, high-throughput sequencing. The on-board processing unit is known as a 'basecalling accelerator' and comprises multiple Graphics Processor Units (GPUs) that combine to provide superior workflow and management of the trace to basecalled data pipeline. The Solid-State Device (SSD) stores 28 Terabytes of data, which facilitates data management and ensures data is not lost if the server is disconnected. The data must be transferred onto a network (i.e. server) for long term storage.
For details on the ports and connections please see the section: Ports and connections
The Data Acquisition Unit dimensions (height x width x depth) are 462 x 178 x 673 mm, and the weight is 28 kg. For power requirements, see "The Power Supply Unit (PSU) and power cables".
The Sequencing Unit
The Sequencing Unit comprises six sequencing strips with eight positions for PromethION flow cells (single port flow cells, namely FLO-PR002) per unit i.e. 48 positions. 24 of the allotted positions (strips 1, 3 and 5) are active. As a result of the use of the FLO-PR002, the sequencing chemistry is the same as other Oxford Nanopore Technologies' platforms, yet the flow cell is engineered differently. The Sequencing Units also contains thermal controls i.e. fans, as well as bespoke electronics that operate the flow cells.
The dimensions (height x width x depth) of the Sequencing Unit are 220 x 437x 410 mm, and the unit weighs 40 kg. For power requirements, see The Power Supply Unit (PSU) and power cables".
The Power Supply Unit (PSU) and power cables
- 2 x 2 kW power supply for the Data Acquisition Unit
- 500 W power supply for the Sequencing Unit
The plugs of the power cables are appropriate for the country of use e.g. NEMA 5-15 grounded (Type B) for United States or BS 1363 (Type G) for the UK. The adaptor to the Data Acquisition Unit or Sequencing Unit are standard three pin female C13 plugs.
For information relating to Power Supply Unit specifications, please see the [PromethION power requirements] (/document/promethion-installation) section.
The PromethION and accessories: itemised
- 48 x Configuration Test Cells
- 3 x Mains to Power Supply cables
- 2 x Data Acquisition Unit to Sequencing Unit PCIe cables
- 1 x Sequencing Unit PSU to Sequencing Unit cable
- 1 x Sequencing Unit Power Supply Unit (PSU)
- 3 x Data Acquisition Unit to Sequencing Unit USB cables
- The PromethION device - 1 x Data Acquisition Unit and 1 x Sequencing Unit

Figure 2. The itemised components present in a PromethION purchase.
PromethION flow cell and Configuration Test Cell
The flow cells or the Configuration Test Cells (CTCs) occupy positions 1 to 24 in strips 1, 3 and 5. The flow cell and CTC details are described in the flow cell section and CTC section.
3. The software
Software overview
PromethION contains all the functional elements of the current Oxford Nanopore Technologies sequencing devices that use GPUs. Below are the components, local or cloud-based, that make up the functional software system for PromethION.
- MinKNOW - Core components and GUI
- Guppy - Basecaller
- EPI2ME - Bioinformatics pipeline
MinKNOW for PromethION
MinKNOW™ Core performs:
- Data acquisition: trace data from current flow at the cathode is collected by the software.
- Real-time analysis and feedback: the trace data is initially processed by MinKNOW before being sent to Guppy for basecalling. Furthermore, strand occupancy of the pores is analysed, presented and fed back to the software and system, reciprocally.
- Data streaming - trace data migrates between MinKNOW and the basecaller assisted by the MinKNOW Core.
- Ensuring chemistry is performing correctly - the pore occupancy measurements provide essential information about the sequencing reactions - the various states e.g. recovery, inactive, sequencing, open, all provide information on individual pores for single molecule interactions with polynucleotides.
MinKNOW GUI performs:
- Device control, including run parameter selection - the MinKNOW GUI can load pre-configured scripts that control parameters such as temperature, voltage, etc. of the real-time sequencing experiment.
- Sample identification and tracking - MinKNOW tracks and saves the current signatures as .fast5 documents, which are migrated to Guppy for basecalling. The MinKNOW GUI allows the appropriate labels to be given to the corresponding sample files, by naming files within a parent folder (see Dogfish for co-ordination of sample .fast5 files).
MinKNOW is regularly updated by Oxford Nanopore Technologies and it continuously improves the MinKNOW core and the GUI to make it more intuitive and keep pace with other technology progress.
Guppy for PromethION
Guppy is the basecaller on PromethION, and it relies on a Recurrent Neural Network to interpret trace current data into a consensus base sequence. Basecalling is the process of converting the electrical signals generated by a DNA or RNA strand passing through the nanopore into the corresponding base sequence of the strand. The general data flow in a nanopore sequencing experiment is shown below.

Raw data - a direct measurement of the changes in ionic current as a DNA/RNA strand passes through the pore. These measurements are recorded by the MinKNOW software. MinKNOW also processes the signal into reads, each read corresponding to a single strand of DNA/RNA.
Basecalling - the raw signal is further processed by the basecalling algorithm to generate the base sequence of the read. The basecaller generates both a .FASTQ and a .fast5 as a default.
Basecalling can be carried out live during the experiment, integrated with data acquisition, in parallel to the experiments, as post-processing after an experiment has finished, or a combination of these.
Sources of basecalling error
There are two main errors an event detection algorithm can make:
- Insertions: when an extra base is inserted where there should not be one - typically, this means a span of raw data points that corresponds to a single set of pore contents.
- Deletions: when a base is missed out.
EPI2ME
EPI2ME™ is a cloud-based platform for bioinformatic data analysis. It is owned by Oxford Nanopore Technologies' subsidiary company, Metrichor™, and thus continues the pipeline for basecalled genomic information from nanopore sequenced polynucleotides. It has an array of workflows, appropriate to the type and preparation of the sample, such that the polynucleotide sequences can be discerned and analysed through the appropriate EPI2ME algorithms.
For a more in depth description of EPI2ME, see the Technical Document.
4. An overview of how it works
An overview of how it works
PromethION is a combination of three modules: the hardware, the software and the flow cells. To achieve successful and high-throughput sequence data, optimised use of kits and reagents is necessary too.
The flow cell obtains the raw current trace data of polynucleotides entering and being sequenced by individual pores; this data is manipulated by the hardware, namely the GPUs, which run the software for basecalling and further downstream analysis. In addition, the hardware configures and maintains accuracy of temperature, efficient running of the sequencing assays, alignment of processing through software and generalised organisation of data trafficking and computer power.
For an in-depth description of the component parts of the hardware, please see: The hardware.
For the software description and how it combines together to perform an information pipeline, please see: The software.
For a description of the flow cell and CTC, please see below sections: The flow cell and The CTC.
5. The flow cell
Overview of the PromethION flow cell
The flow cell contains the active molecules and ions of the sequencing chemistry, which enables polynucleotides to be resolved at the individual base level; a discrete base sequence is generated as a result of the prepared library being added to the flow cell and manipulated by the software package on PromethION.
The flow cell contains up to 12,000 pores in individual wells; 3000 pores can be actively sequencing at one time, i.e. can be measured by the flow cell Application Specific Integrated Circuitry (ASIC). This is called multiplexing (MUX), where four pores, interact with a signal amplifier on the ASIC. This is important for MUX pore group selection during the sequencing experiment and is fundamental to high throughput and reliable quality reads. The quality of the pore-membrane complex is a factor in the MUX scan group qualification, such that pores from sequencing MUXs (e.g. wells with single-pore complex in) are entered into Group 1, then distributed in decreasing amounts into Group 2, Group 3 and Group 4.
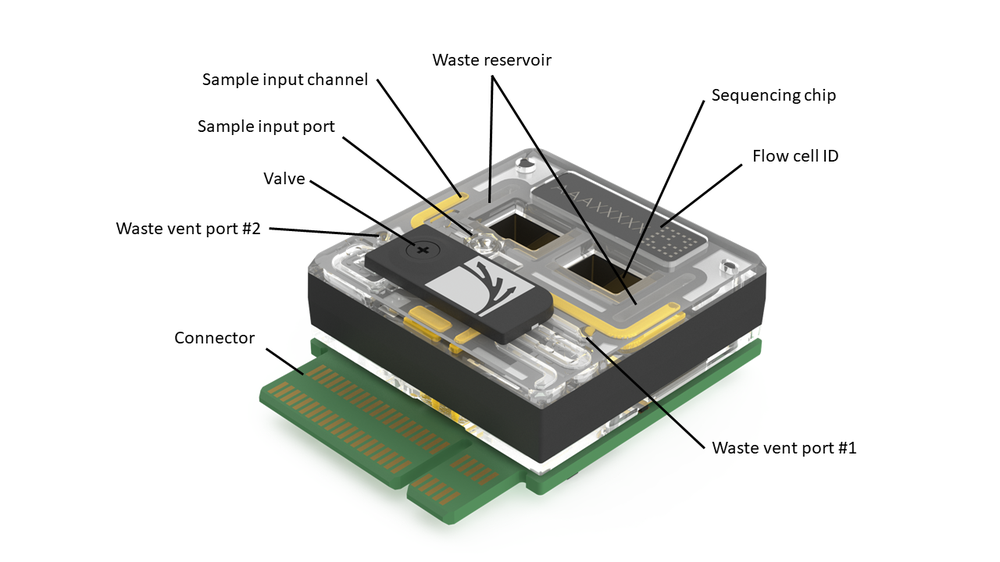
Figure 3. The single-port flow cell FLO-PRO002. The FLO-PRO002 is shown in the above figure. It is shown in the open state i.e. during sample loading. The sample flows over the chip to allow sequencing of the sample's content polynucleotides.
A flow cell contains a sample reservoir directly above the sequencing array. Excess fluid passes out of the chambers into through the waste reservoir and can be siphoned from one of the waste ports only when the valve is closed. A rotating valve is present over the input ports and should be closed during an experiment to prevent sample leakage.
The PromethION flow cell physics and chemistry
For a detailed explanation of the biology, chemistry and physics of the flow cell, please refer to the following Technical Document
6. The Configuration Test Cell
The PromethION Configuration Test Cell (CTC)
Configuration is the process of testing the communication between the PromethION modules and the control software on the PromethION computer, and that it is operational prior to experimental work being performed. This is carried out in the absence of any chemistry and uses a specific flow cell known as the Configuration Test Cell (CTC). The CTC contains the same circuit board as that found on PromethION flow cells, and is inserted into the sensing module of the PromethION.


Figure 4. The Configuration Test Cell.
7. An overview of what is needed
An overview of what is needed
PromethION is a much larger, higher throughput device than other Oxford Nanopore Technologies devices. As a result, some basic facility requirements are needed for suitable running of the PromtheION; these are described below.
PromethION can deliver several Terabytes of data within a 64-hour run time; the internal memory of the PromethION is intended to act as a storage buffer only. Therefore to ensure confluency in data acquisition, both fast data trafficking technology (10 Gbps) and high capacity (10s of Terabytes) storage servers are essential for efficient running and data collection from a PromethION.
In addition to these specifications, general laboratory integration is necessary and as such, requirements like a stable physical support (for the 40 kg Sequencing Unit) and peripherals such as a mouse and keyboard, are needed.
The details of the facility installation requirements are outlined in the section: Infrastructure.
8. Infrastructure
Essential requirements - connections and ports
Below are the details of the necessary connections required for data transfer, IT integration and powering both units of the PromethION. The connections are the interaction of the PromethION within an IT system that, including the PromethION, allows data extraction from a biological sample and thus allows manipulation by IT infrastructure i.e. as .fast5 and FASTQ files on a personal computer.
The labelled ports are absolutely necessary for PromethION function. The ports screened off with a yellow 'not permitted' sign (below) indicate that these ports are superfluous and not used by Oxford Nanopore Technologies or any user.

The rear Power and Networking panel of the Data Acquisition Unit module has the following ports:

The rear Power and Networking panel of the Sequencing Unit module has the following ports:
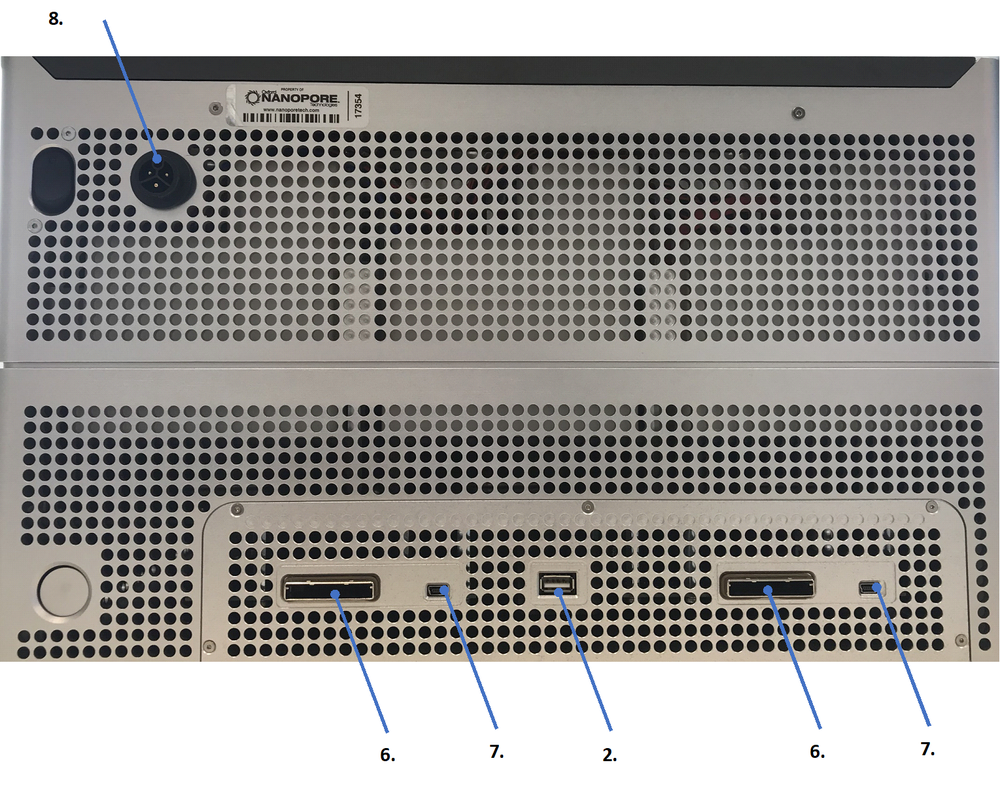
The connections and ports are listed in Table 1. Data Acquisition Unit, DAU; Sequencing Unit, SQU.
| Port Label | Unit | Copper | Fibre Optics |
|---|---|---|---|
| 1. | DAU | a. 2 x 10 Gbps RJ45 ethernet adapters (+) | b. 2 x 10 Gbps with DCHP capability + 2 x SFP+ modules |
| 1. | DAU | a. 2 x 10 Gbps Copper cables (-) | b. 2 x 10 Gbps fibre optic cables |
| 2. | DAU | USB-USB port and cable (.) | |
| 3. | DAU | Spare USB port or for USB mouse or USB keyboard (=) | |
| 4. | DAU | VGA monitor (#) | |
| 5. | DAU | 2 x 2kW C-13 power port and cable for DAU | |
| 6. | SQU/DAU | 2 x PCIe adapter and cables (..) | |
| 7. | SQU/DAU | 2 x mini USB-USB port and cables (.) | |
| 8. | SQU | 500 W C-13 Power port and cable for SQU | |
(+) The lower shielded RJ45 port is used for initial address configuration. Dual port setup can be managed for data transfer once configured via the lower port (as indicated in the installation guide). (-) Optimally, shielded high capacity (10 Gbps) cables should be used i.e. Cat5e. (=) The 1 x USB port at the back or the 2 x USB ports on the front of the Data Acquisition Unit can be used for the mouse and keyboard. (#) The monitor will require a C-13 power cable; this cable will not plug into anywhere on the Data Acquisition Unit to power the monitor. (.) This USB-USB and mini-USB cables are used for firmware upgrades. (..) The PCIe cables link the Sequencing Unit/Data Acquisition Unit for data transfer. The PCIe cards function only as a combination of a single card and one of a pair card; thus, one port remains vacant.
9. Safety
Manufacturing information
The PromethION Alpha-Beta is made by: Oxford Nanopore Technologies Robert Robinson Avenue Oxford Science Park OX4 4GA United Kingdom.
The product is protected by the patents and patents pending in the website below: https://nanoporetech.com/patents
Intended use
Oxford Nanopore Technologies® PromethION™ device is an electronic analysis system for use in scientific research. The core technology is built around a nanopore that is able to detect single molecule events of nucleic acids (DNA/RNA).
It is for research use only and not for in vitro diagnostic or therapeutic use.
The purpose of the PromethION safety information
The safety information provides you with the details needed to install and use the system safely.
Electrical information
Sequencing Unit
| Parameter | Values |
|---|---|
| Supply voltage | 100-240 V (50/60 Hz) |
| Input current | 5.5 - 2.7 A |
| Maximum power | 500 W |
| Maximum PSU output current | 21 A |
| Maximum PSU output voltage | 24 VDC |
Data Acquisition Unit
| Parameter | Values |
|---|---|
| Supply voltage | 100-240 V (50/60 Hz) |
| Maximum power | 2200 W |
| Maximum current | 12-11 A (220-240 V - 2200 W) |
| Maximum current | 14-11 A (100-127 V - 1200 W) |
Technical Specification
Sequencing Unit
| Component Specification | Details |
|---|---|
| Size and weight | H 220 x W 437 x D 410 mm, 40 kg |
| Sequencing spec: | 48 sequencing positions for flow cells |
| Thermal Management | Designed to sequence at room temperature (+18-25° C). Users should allow 30 cm clearance to the rear and sides of the device. CAUTION: Rear of instrument heats up during operation |
Data Acquisition Unit
| Component Specification | Details |
|---|---|
| Size and weight | H 462 x W 178 x D 673 mm, 28 kg |
| Compute spec | 32 TB SSD Storage, 384 GB RAM, Intel CPU, PromethION Accelerator |
| Pre-loaded software | Ubuntu OS, PromethION OS (MinKNOW inside) |
| Thermal Management | Designed to sequence at room temperature (+18-25° C) Users should allow 30 cm clearance to the rear and sides of the device CAUTION: Rear of instrument heats up during operation |
Connections to the device

- C13 power cables
- USB ports
- RJ45 Copper Ethernet ports
- VGA port
- SFP+ fibre modules
- PCIe cable ports
Safety notices

Warning - WARNING indicates a hazardous situation which, if not avoided, could result in death or serious injury. It is important not to proceed until all stated conditions are met and clearly understood.

Caution - CAUTION indicates a hazardous situation which, if not avoided, could result in minor or moderate injury. It is important not to proceed until all stated conditions are met and clearly understood.

Advisory - ADVISORY indicates instructions that must be followed to avoid damage to the product or other equipment.
Note - A Note is used to indicate information that is important for trouble-free and optimal use of the product.
Critical step - A Critical step is used to indicate information that is essential for trouble-free and optimal use of the product.
Tip - A Tip contains useful information that could improve or optimise your procedure.
The safety notices below are intended to supplement, not supersede, the normal safety requirements prevailing in the user’s country.
Material Safety Data Sheets (MSDSs)

- The MSDSs for any chemicals supplied by Oxford Nanopore Technologies are available on request to support@nanoporetech.com.

- Read and understand the MSDSs before handling, working with or storing the chemicals being used within the Oxford Nanopore Technologies® sequencing device.

- Minimise contact with the chemicals by wearing protective clothing, safety glasses and gloves. The MSDS will carry specific requirements.

- Minimise inhalation of chemicals by using appropriate and adequate ventilation. The MSDS will carry any specific requirements. Continuously check for any spills or leakages. If a spill or leak occurs, follow the clean-up guidelines provided on the MSDS.

- All components of the sequencing device should be handled, stored and disposed of in accordance with local, state/provincial or national laws and regulations.
General precautions

- When handling toxic, radioactive or pathogenic samples as defined by the WHO Laboratory Biosafety Manual, observe the safety regulations of the specific local in question.

- Do not use the sequencing device if it has suffered any damage, e.g. to power cables, data transfer cables, power supplies or flow cells.
Personal protection

- Specimens and reagents containing materials from humans should be treated as potentially infectious. Use safe laboratory procedures as outlined in publications such as Biosafety in Microbiological and Biomedical Laboratories (http://www.cdc.gov/biosafety/publications/bmbl5/index.htm).

- The operator has to take all necessary actions to avoid spreading hazardous biological agents in the vicinity of the system. The facility should comply with the national code of practice for biosafety.

- Samples being loaded into the flow cell should be used, stored and disposed of according to the required safety regulations and laws. Consult the responsible body for safety in your lab for local regulations.

- Samples containing infectious agents should be handled with the greatest of care and in accordance with the required safety regulations and laws.

- It is good laboratory practice to always wear safety glasses, gloves (2 pairs if working with infectious agents) and a lab coat. There may be other locally advised items which add to this recommendation.
Use of the PromethION, flow cells and reagents

- When handling toxic, radioactive or pathogenic samples as defined by the WHO Laboratory Biosafety Manual, observe the specific local regulations.

- Loading excess buffer, sample or de-ionised water to the flow cell will cause an overflow of the waste compartment. Absorbent material should be used to capture sample and buffer which will come out through the waste port. All material should be disposed of in line with local regulations for biological waste.

- In the unlikely event that the sequencing device is found to be hot during use, terminate use of the device from the operating computer and refer to troubleshooting guidelines.
Maintenance

Repairs must only be performed by Oxford Nanopore Technologies, and no components should be replaced. Contact support@nanoporetech.com in the event of damage to the sequencing device or the flow cells.
Before using cleaning or decontamination methods other than those stipulated by the manufacturer, contact the manufacturer to ensure that the intended method will not damage the sequencing device and/or flow cell.
When returning sequencing devices and/or flow cells, ensure that they are fully decontaminated and do not present any kind of health risk to our staff.
Disposal and recycling instructions

- Used plasticware, such as reagents, tubes and pipette tips, must be collected and disposed of properly in accordance with local safety regulations and laboratory procedures.

- The flow cell buffer, the sample preparation kit buffers and wash kit buffers must not be mixed and must be kept away from strong acids and alkalis.

- The flow cell buffer, wash kit buffers and sample preparation kit buffers must be disposed of according to the local regulations. They must not be disposed of down a sink.

- The Terms and Conditions for the use of sequencing devices stipulate any flow cells that have been used with or otherwise been in contact with materials of Biohazard Level 3 or higher (“Contaminated Flow Cells”) must not be returned. Proof of legal and appropriate destruction of any Contaminated Flow cells will be required.

- The sequencing device shall be decontaminated before decommissioning, and all local regulations for electronic and electrical waste shall be followed with regard to disposal of the components if they are not being returned to Oxford Nanopore Technologies.

- The flow cell shall be disposed of as hazardous biological waste, and all local regulation for such waste shall be followed if they are not being returned to Oxford Nanopore Technologies.
Labels on the instrument
Labels on the PromethION Alpha-Beta Sequencing Unit:


Labels on the PromethION Alpha-Beta Data Acquisition Unit:


Label on the flow cell:

Emergency procedures
In case of emergency, switch the PromethION off at the power switch and unplug the power cable from the back of the device.






