Rapid sequencing gDNA - barcoding (Q-SQK-RBK110.96) (Q_RBK_revB_26Sep2024)
GridION: Protocol
V Q_RBK_revB_26Sep2024
FOR RESEARCH USE ONLY
Contents
Introduction to the protocol
Library preparation
Sequencing and data analysis
1. Overview of the protocol
Rapid Barcoding Kit features
This kit is recommended for users who:
- Wish to multiplex samples to reduce price per sample
- Need a PCR-free method of multiplexing to preserve additional information such as base modifications
- Require a short preparation time
- Have limited access to laboratory equipment
Introduction to the Rapid Barcoding Kit 96
This protocol describes how to carry out rapid barcoding of genomic DNA using the Rapid Barcoding Kit 96 (Q-SQK-RBK110.96).
Steps in the sequencing workflow:
Prepare for your experiment
You will need to:
- Extract your DNA and check its length, quantity, and purity. The quality checks performed on input samples are essential in ensuring experimental success.
- Ensure you have the Q-Line Rapid Barcoding Kit 96, the correct equipment, and third-party reagents
Library preparation
You will need to:
- Tagment your DNA using the Rapid Barcodes in the kit; this simultaneously attaches a pair of barcodes to the fragments
- Pool the barcoded samples
- Attach sequencing adapters supplied in the kit to the DNA ends
- Prime the flow cell and load your DNA library into the flow cell
Sequencing and analysis
You will need to:
- Start an assay using the sequencing software, which will collect raw data from the device and convert it into basecalled reads. There is also an option to demultiplex reads into barcode-specific folders.
Compatibility of this protocol
This protocol should only be used in combination with:
- Rapid Barcoding Kit 96 (Q-SQK-RBK110.96)
- R9.4.1 flow cells (Q-FLO-MIN106D)
- Flow Cell Wash Kit (Q-EXP-WSH004)
2. Equipment and consumables
Material
- 50 ng high molecular weight genomic DNA per sample
- Rapid Barcoding Kit 96 (Q-SQK-RBK110.96)
Consumibles
- 1.5 ml Eppendorf DNA LoBind tubes
- Tubos Eppendorf DNA LoBind de 2 ml
- 0.2 ml thin-walled PCR tubes
- Placa de PCR Eppendorf twin.tec® LoBind de 96 pocillos, con semifaldón y sellado térmico (Eppendorf™, 0030129504)
- Nuclease-free water (e.g. Thermo Scientific, AM9937)
- Etanol al 80 % recién preparado con agua sin nucleasas
Instrumental
- Ice bucket with ice
- Centrifugadora de microplacas
- Timer
- Termociclador o termobloque
- Gradilla magnética
- Hula mixer (gentle rotator mixer)
- P1000 pipette and tips
- P200 pipette and tips
- P100 pipette and tips
- P20 pipette and tips
- P2 pipette and tips
- Multichannel pipette and tips
Equipo opcional
- Standard gel electrophoresis equipment
- Bioanalizador Agilent (o equivalente)
- Qubit™ fluorometer (or equivalent for QC check)
For this protocol, you will need 50 ng high molecular weight genomic DNA per sample.
Input DNA
How to QC your input DNA
It is important that the input DNA meets the quantity and quality requirements. Using too little or too much DNA, or DNA of poor quality (e.g. highly fragmented or containing RNA or chemical contaminants) can affect your library preparation.
For instructions on how to perform quality control of your DNA sample, please read the Input DNA/RNA QC protocol.
Chemical contaminants
Depending on how the DNA is extracted from the raw sample, certain chemical contaminants may remain in the purified DNA, which can affect library preparation efficiency and sequencing quality. Read more about contaminants on the Contaminants page of the Community.
Rapid Barcoding Kit 96 (Q-SQK-RBK110.96) contents
| Name | Acronym | Cap colour | No. of vials | Fill volume per vial (µl) |
|---|---|---|---|---|
| Rapid Barcode plate | RB96 | - | 3 plates | 8 µl per well |
| AMPure XP Beads | AXP | Brown | 3 | 1,200 |
| Sequencing Buffer II | SBII | Red | 1 | 500 |
| Rapid Adapter F | RAP-F | Green | 1 | 25 |
| Elution Buffer | EB | Black | 1 | 500 |
| Loading Beads II | LBII | Pink | 1 | 360 |
| Loading Solution | LS | White cap, pink label | 1 | 400 |
| Flush Tether | FLT | Purple | 1 | 400 |
| Flush Buffer | FB | White | 1 bottle | 15,500 |
This product contains AMPure XP reagent manufactured by Beckman Coulter, Inc.
Rapid barcode sequences
| Component | Sequence |
|---|---|
| RB01 | AAGAAAGTTGTCGGTGTCTTTGTG |
| RB02 | TCGATTCCGTTTGTAGTCGTCTGT |
| RB03 | GAGTCTTGTGTCCCAGTTACCAGG |
| RB04 | TTCGGATTCTATCGTGTTTCCCTA |
| RB05 | CTTGTCCAGGGTTTGTGTAACCTT |
| RB06 | TTCTCGCAAAGGCAGAAAGTAGTC |
| RB07 | GTGTTACCGTGGGAATGAATCCTT |
| RB08 | TTCAGGGAACAAACCAAGTTACGT |
| RB09 | AACTAGGCACAGCGAGTCTTGGTT |
| RB10 | AAGCGTTGAAACCTTTGTCCTCTC |
| RB11 | GTTTCATCTATCGGAGGGAATGGA |
| RB12 | CAGGTAGAAAGAAGCAGAATCGGA |
| RB13 | AGAACGACTTCCATACTCGTGTGA |
| RB14 | AACGAGTCTCTTGGGACCCATAGA |
| RB15 | AGGTCTACCTCGCTAACACCACTG |
| RB16 | CGTCAACTGACAGTGGTTCGTACT |
| RB17 | ACCCTCCAGGAAAGTACCTCTGAT |
| RB18 | CCAAACCCAACAACCTAGATAGGC |
| RB19 | GTTCCTCGTGCAGTGTCAAGAGAT |
| RB20 | TTGCGTCCTGTTACGAGAACTCAT |
| RB21 | GAGCCTCTCATTGTCCGTTCTCTA |
| RB22 | ACCACTGCCATGTATCAAAGTACG |
| RB23 | CTTACTACCCAGTGAACCTCCTCG |
| RB24 | GCATAGTTCTGCATGATGGGTTAG |
| RB25 | GTAAGTTGGGTATGCAACGCAATG |
| RB26 | CATACAGCGACTACGCATTCTCAT |
| RB27 | CGACGGTTAGATTCACCTCTTACA |
| RB28 | TGAAACCTAAGAAGGCACCGTATC |
| RB29 | CTAGACACCTTGGGTTGACAGACC |
| RB30 | TCAGTGAGGATCTACTTCGACCCA |
| RB31 | TGCGTACAGCAATCAGTTACATTG |
| RB32 | CCAGTAGAAGTCCGACAACGTCAT |
| RB33 | CAGACTTGGTACGGTTGGGTAACT |
| RB34 | GGACGAAGAACTCAAGTCAAAGGC |
| RB35 | CTACTTACGAAGCTGAGGGACTGC |
| RB36 | ATGTCCCAGTTAGAGGAGGAAACA |
| RB37 | GCTTGCGATTGATGCTTAGTATCA |
| RB38 | ACCACAGGAGGACGATACAGAGAA |
| RB39 | CCACAGTGTCAACTAGAGCCTCTC |
| RB40 | TAGTTTGGATGACCAAGGATAGCC |
| RB41 | GGAGTTCGTCCAGAGAAGTACACG |
| RB42 | CTACGTGTAAGGCATACCTGCCAG |
| RB43 | CTTTCGTTGTTGACTCGACGGTAG |
| RB44 | AGTAGAAAGGGTTCCTTCCCACTC |
| RB45 | GATCCAACAGAGATGCCTTCAGTG |
| RB46 | GCTGTGTTCCACTTCATTCTCCTG |
| RB47 | GTGCAACTTTCCCACAGGTAGTTC |
| RB48 | CATCTGGAACGTGGTACACCTGTA |
| RB49 | ACTGGTGCAGCTTTGAACATCTAG |
| RB50 | ATGGACTTTGGTAACTTCCTGCGT |
| RB51 | GTTGAATGAGCCTACTGGGTCCTC |
| RB52 | TGAGAGACAAGATTGTTCGTGGAC |
| RB53 | AGATTCAGACCGTCTCATGCAAAG |
| RB54 | CAAGAGCTTTGACTAAGGAGCATG |
| RB55 | TGGAAGATGAGACCCTGATCTACG |
| RB56 | TCACTACTCAACAGGTGGCATGAA |
| RB57 | GCTAGGTCAATCTCCTTCGGAAGT |
| RB58 | CAGGTTACTCCTCCGTGAGTCTGA |
| RB59 | TCAATCAAGAAGGGAAAGCAAGGT |
| RB60 | CATGTTCAACCAAGGCTTCTATGG |
| RB61 | AGAGGGTACTATGTGCCTCAGCAC |
| RB62 | CACCCACACTTACTTCAGGACGTA |
| RB63 | TTCTGAAGTTCCTGGGTCTTGAAC |
| RB64 | GACAGACACCGTTCATCGACTTTC |
| RB65 | TTCTCAGTCTTCCTCCAGACAAGG |
| RB66 | CCGATCCTTGTGGCTTCTAACTTC |
| RB67 | GTTTGTCATACTCGTGTGCTCACC |
| RB68 | GAATCTAAGCAAACACGAAGGTGG |
| RB69 | TACAGTCCGAGCCTCATGTGATCT |
| RB70 | ACCGAGATCCTACGAATGGAGTGT |
| RB71 | CCTGGGAGCATCAGGTAGTAACAG |
| RB72 | TAGCTGACTGTCTTCCATACCGAC |
| RB73 | AAGAAACAGGATGACAGAACCCTC |
| RB74 | TACAAGCATCCCAACACTTCCACT |
| RB75 | GACCATTGTGATGAACCCTGTTGT |
| RB76 | ATGCTTGTTACATCAACCCTGGAC |
| RB77 | CGACCTGTTTCTCAGGGATACAAC |
| RB78 | AACAACCGAACCTTTGAATCAGAA |
| RB79 | TCTCGGAGATAGTTCTCACTGCTG |
| RB80 | CGGATGAACATAGGATAGCGATTC |
| RB81 | CCTCATCTTGTGAAGTTGTTTCGG |
| RB82 | ACGGTATGTCGAGTTCCAGGACTA |
| RB83 | TGGCTTGATCTAGGTAAGGTCGAA |
| RB84 | GTAGTGGACCTAGAACCTGTGCCA |
| RB85 | AACGGAGGAGTTAGTTGGATGATC |
| RB86 | AGGTGATCCCAACAAGCGTAAGTA |
| RB87 | TACATGCTCCTGTTGTTAGGGAGG |
| RB88 | TCTTCTACTACCGATCCGAAGCAG |
| RB89 | ACAGCATCAATGTTTGGCTAGTTG |
| RB90 | GATGTAGAGGGTACGGTTTGAGGC |
| RB91 | GGCTCCATAGGAACTCACGCTACT |
| RB92 | TTGTGAGTGGAAAGATACAGGACC |
| RB93 | AGTTTCCATCACTTCAGACTTGGG |
| RB94 | GATTGTCCTCAAACTGCCACCTAC |
| RB95 | CCTGTCTGGAAGAAGAATGGACTT |
| RB96 | CTGAACGGTCATAGAGTCCACCAT |
3. Computer requirements and software
GridION IT requirements
The GridION device contains all the hardware required to control up to five sequencing experiments simultaneously and acquire the data. The device is further enhanced with high performance GPU technology for real-time basecalling. Read more in the GridION Q IT Requirements document in the Q system channel.
The sequencing software
The sequencing software controls the GridION, collects sequencing data in real-time and processes it into basecalls. The software can also demultiplex reads by barcode, and basecall/demultiplex data after a sequencing run has completed. You will be using the sequencing software for every assay you run.
For instructions on how to run the sequencing software on the GridION, please refer to the Q-Line sequencing software user guide.
4. Library preparation
Material
- 50 ng high molecular weight genomic DNA per sample
- Rapid Barcode Plate (RB96)
- Rapid Adapter F (RAP F)
- AMPure XP Beads (AXP, or SPRI)
- Elution Buffer (EB)
Consumibles
- 0.2 ml thin-walled PCR tubes
- Placa de PCR Eppendorf twin.tec® LoBind de 96 pocillos, con semifaldón y sellado térmico (Eppendorf™, 0030129504)
- 1.5 ml Eppendorf DNA LoBind tubes
- Tubos Eppendorf DNA LoBind de 2 ml
- Nuclease-free water (e.g. Thermo Scientific, AM9937)
- Etanol al 80 % recién preparado con agua sin nucleasas
Instrumental
- Ice bucket with ice
- Timer
- Thermal cycler
- Centrifugadora de microplacas
- Gradilla magnética
- Hula mixer (gentle rotator mixer)
- P1000 pipette and tips
- P200 pipette and tips
- P100 pipette and tips
- P20 pipette and tips
- P10 pipette and tips
- P2 pipette and tips
- Multichannel pipette and tips
Equipo opcional
- Standard gel electrophoresis equipment
- Bioanalizador Agilent (o equivalente)
- Qubit™ fluorometer (or equivalent for QC check)
Program the thermal cycler: 30°C for 2 minutes, then 80°C for 2 minutes.
Thaw kit components at room temperature, spin down briefly using a microfuge and mix by pipetting as indicated by the table below:
| Reagent | 1. Thaw at room temperature | 2. Briefly spin down | 3. Mix well by pipetting |
|---|---|---|---|
| Rapid Barcode plate (RB96) | Not frozen | ✓ | ✓ |
| Rapid Adapter F (RAP-F) | Not frozen | ✓ | ✓ |
| AMPure XP Beads (AXP, or SPRI) | ✓ | ✓ | Mix by pipetting or vortexing immediately before use |
| Sequencing Buffer II (SBII) | ✓ | ✓ | ✓* |
| Loading Beads II (LBII) | ✓ | ✓ | Mix by pipetting or vortexing immediately before use |
| Elution Buffer (EB) | ✓ | ✓ | ✓ |
| Flush Buffer (FB) | ✓ | ✓ | Mix by vortexing |
| Flush Tether (FLT) | ✓ | ✓ | ✓ |
*Vortexing, followed by a brief spin in a microfuge, is recommended for Sequencing Buffer II (SBII) and Flush Buffer (FB).
Prepare the DNA in nuclease-free water.
- Transfer 50 ng genomic DNA per sample into a 1.5 ml Eppendorf DNA LoBind tube
- Adjust the volume to 9 μl with nuclease-free water
- Mix by pipetting
- Spin down briefly in a microfuge
In 0.2 ml thin-walled PCR tubes or an Eppendorf twin.tec® PCR plate 96 LoBind, mix the following. The Rapid Barcodes can be transferred using a multichannel pipette:
| Reagent | Volume |
|---|---|
| 50 ng template DNA | 9 μl |
| Rapid Barcodes (RB01-96, one for each sample) | 1 μl |
| Total | 10 μl |
Ensure the components are thoroughly mixed by pipetting and spin down briefly.
Incubate the tubes or plate at 30°C for 2 minutes and then at 80°C for 2 minutes. Briefly put the tubes or plate on ice to cool.
Pool all barcoded samples, noting the total volume.
Resuspend the AMPure XP Beads (AXP) by vortexing.
To the entire pooled barcoded sample from Step 7, add an equal volume of resuspended AMPure XP Beads (AXP) and mix by flicking the tube.
Incubate on a Hula mixer (rotator mixer) for 5 minutes at room temperature.
Prepare at least 3 ml of fresh 80% ethanol in nuclease-free water.
Spin down the sample and pellet on a magnet. Keep the tube on the magnet, and pipette off the supernatant.
Keep the tube on the magnet and wash the beads with 1.5 ml of freshly prepared 80% ethanol without disturbing the pellet. Remove the ethanol using a pipette and discard.
Repeat the previous step.
Briefly spin down and place the tube back on the magnet. Pipette off any residual ethanol. Allow to dry for 30 seconds, but do not dry the pellet to the point of cracking.
Remove the tube from the magnetic rack and resuspend the pellet in 15 µl Elution Buffer (EB). Incubate for 10 minutes at room temperature.
Pellet the beads on a magnet until the eluate is clear and colourless, for at least 1 minute.
Remove and retain 15 µl of eluate into a clean 1.5 ml Eppendorf DNA LoBind tube.
- Remove and retain the eluate which contains the DNA library in a clean 1.5 ml Eppendorf DNA LoBind tube
- Dispose of the pelleted beads
Quantify 1 µl of eluted sample using a Qubit fluorometer.
Transfer 11 µl of the sample into a clean 1.5 ml Eppendorf DNA LoBind tube.
Add 1 µl of Rapid Adapter F (RAP F) to 11 µl of barcoded DNA.
Mix gently by flicking the tube, and spin down.
Incubate the reaction for 5 minutes at room temperature.
The prepared library is used for loading into the flow cell. Store the library on ice or at 4°C until ready to load.
5. Priming and loading the MinION Flow Cell
Material
- Flush Tether (FLT)
- Flush Buffer (FB)
- Sequencing Buffer II (SBII)
- Loading Beads II (LBII)
- Loading Solution (LS)
Consumibles
- 1.5 ml Eppendorf DNA LoBind tubes
Instrumental
- GridION device
- P1000 pipette and tips
- P100 pipette and tips
- P20 pipette and tips
Priming and loading a flow cell
We recommend all new users watch the 'Priming and loading your flow cell' video before your first run.
Using the Loading Solution
We recommend using the Loading Beads II (LBII) for loading your library onto the flow cell for most sequencing experiments. However, if you have previously used water to load your library, you must use Loading Solution (LS) instead of water. Note: some customers have noticed that viscous libraries can be loaded more easily when not using Loading Beads II.
Thaw the Sequencing Buffer II (SBII), Loading Beads II (LBII) or Loading Solution (LS, if using), Flush Tether (FLT) and one bottle of Flush Buffer (FB) at room temperature before mixing the reagents by vortexing and spin down at room temperature.
To prepare the flow cell priming mix, add 30 µl of thawed and mixed Flush Tether (FLT) directly to the bottle of thawed and mixed Flush Buffer (FB), and mix by vortexing at room temperature.
Slide open the GridION lid and insert the flow cell.
Press down firmly on the flow cell to ensure correct thermal and electrical contact.
Complete a flow cell check to assess the number of pores available before loading the library.
This step can be omitted if the flow cell has been checked previously.
See the flow cell check instructions in the MinKNOW protocol for more information.
Slide the flow cell priming port cover clockwise to open the priming port.
Take care when drawing back buffer from the flow cell. Do not remove more than 20-30 µl, and make sure that the array of pores are covered by buffer at all times. Introducing air bubbles into the array can irreversibly damage pores.
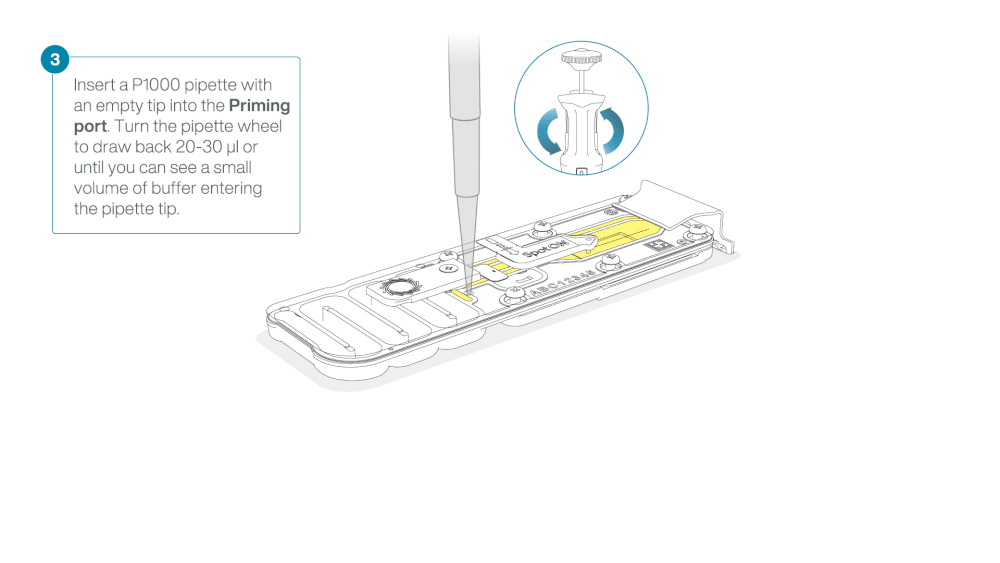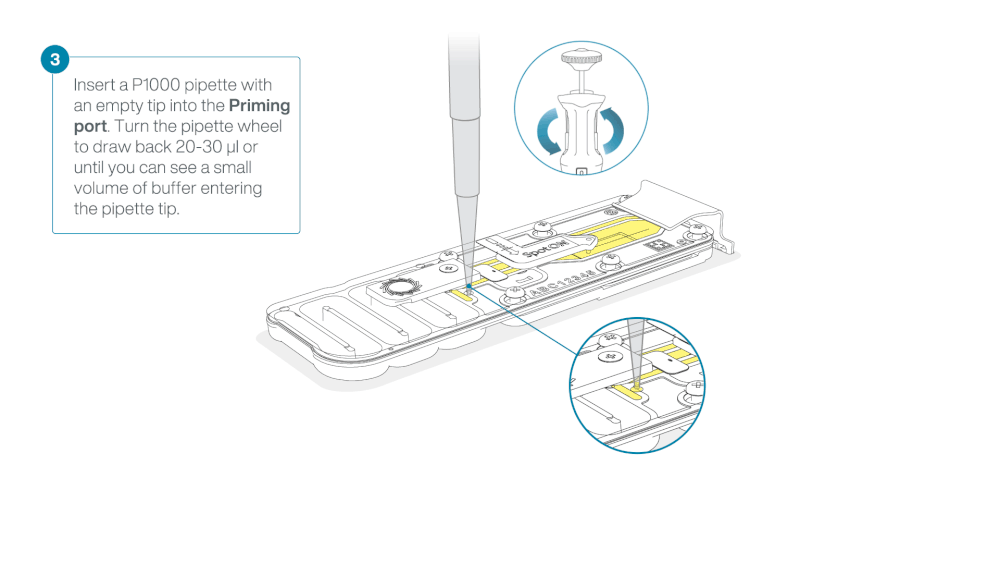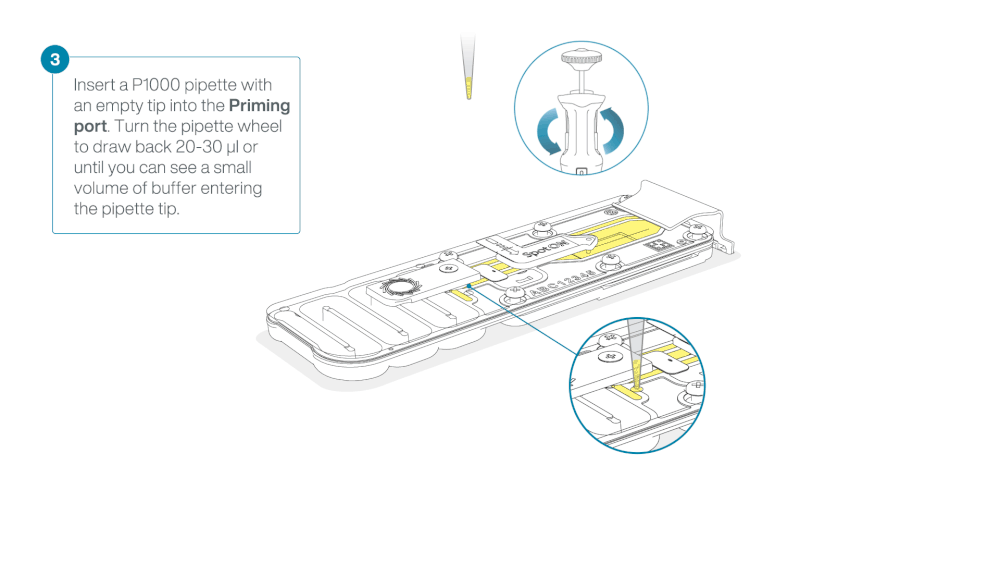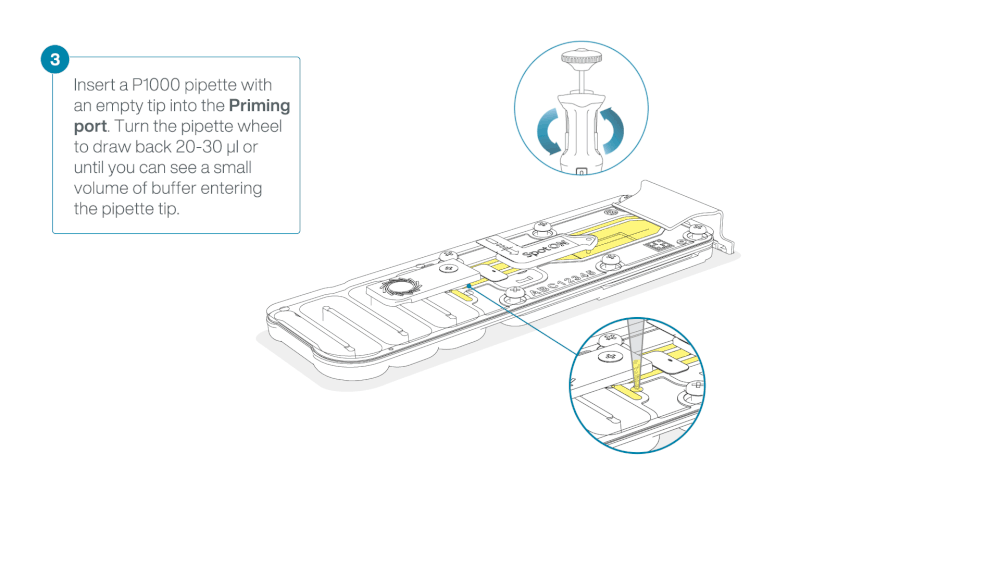
After opening the priming port, check for a small air bubble under the cover. Draw back a small volume to remove any bubbles:
- Set a P1000 pipette to 200 µl
- Insert the tip into the priming port
- Turn the wheel until the dial shows 220-230 µl, to draw back 20-30 µl, or until you can see a small volume of buffer entering the pipette tip
Note: Visually check that there is continuous buffer from the priming port across the sensor array.
Load 800 µl of the priming mix into the flow cell via the priming port, avoiding the introduction of air bubbles. Wait for five minutes. During this time, prepare the library for loading by following the steps below.

Thoroughly mix the contents of the Loading Beads II (LBII) by pipetting.
The Loading Beads II (LBII) tube contains a suspension of beads. These beads settle very quickly. It is vital that they are mixed immediately before use.
In a new tube, prepare the library for loading as follows:
| Reagent | Volume per flow cell |
|---|---|
| Sequencing Buffer II (SBII) | 37.5 µl |
| Loading Beads II (LBII) mixed immediately before use, or Loading Solution (LS), if using | 25.5 µl |
| DNA library | 12 µl |
| Total | 75 µl |
Note: Load the library onto the flow cell immediately after adding the Sequencing Buffer II (SBII) and Loading Beads II (LBII).
Complete the flow cell priming:
- Gently lift the SpotON sample port cover to make the SpotON sample port accessible.
- Load 200 µl of the priming mix into the flow cell priming port (not the SpotON sample port), avoiding the introduction of air bubbles.

Mix the prepared library gently by pipetting up and down just prior to loading.
Add 75 μl of the prepared library to the flow cell via the SpotON sample port in a dropwise fashion. Ensure each drop flows into the port before adding the next.

Gently replace the SpotON sample port cover, making sure the bung enters the SpotON port, close the flow cell priming port and close the GridION lid.
6. Data acquisition and basecalling
How to start sequencing
Follow the instructions in the Q-Line sequencing software user guide beginning from the "Set up and run an assay" section.







